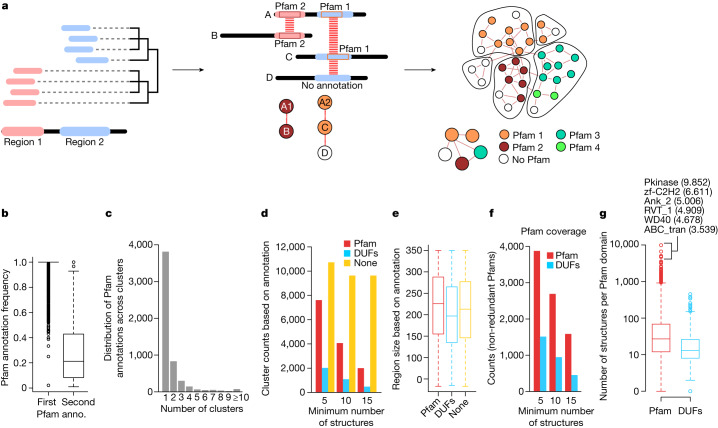
Fig. 4. Prediction of domain families by local structural similarity hits.
a, Diagram of the structure-based domain family prediction method. Clustering of the start and end positions for Foldseek hits of one protein against all others was used to define potential domain boundary positions. Each predicted domain region was linked to the others sharing structural similarities and graph-based clustering was used to define domain families and interdomain similarity. b, The frequency distribution of the most common (n = 9,631) and the second most common (n = 1,628) Pfam annotations found members of all predicted domain families. anno., annotation. c, The counts of the number of clusters with a given Pfam as the most frequent. d, The number of domain family clusters annotated to a Pfam, DUF or no domain annotation. e, The distribution of protein region length in the predicted domain families, stratified by their annotations: Pfam domain (n = 1,048,276), DUF (n = 72,798) and not annotated (n = 1,904,498). f, Non-redundant count of Pfam and DUF domain families found in the structure-based predicted families. g, The distribution of the number of structures found for each predicted domain family annotated with a known Pfam (n = 3,875) or DUF domain (n = 1,513). The top 6 Pfam annotations are highlighted using their abbreviations: Pkinase, protein kinase domain PF00069); zf-C2H2, zinc finger, C2H2 type PF00096; Ank_2, ankyrin repeats, PF12796; RVT_1, reverse transcriptase, PF00078; WD40, WD domain, G-beta repeat PF00400; ABC_tran, ABC transporter, PF00005. The box plots in b, e and g show the median (centre line), the quartiles 1 and 3 (box limits) and 1.5 × the interquartile range (whiskers).