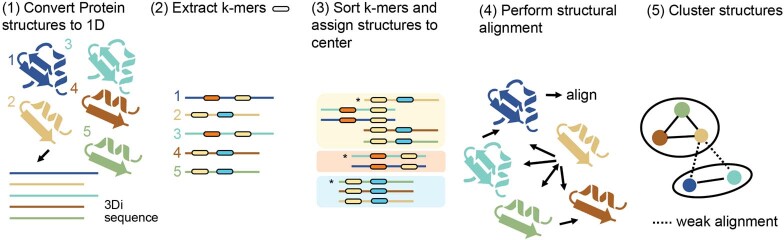
Extended Data Fig. 1. The five-step clustering pipeline for efficiently clustering millions of protein structures using Foldseek’s 3Di alphabet.
(1) Protein structures are converted to 3Di sequences and processed through the Linclust workflow. (2) For each sequence, 300 min-hasing k-mers are extracted and sorted. (3) The longest structure is assigned to be the centre of each k-mer cluster. (4) Structural alignment is performed in two stages: first an ungapped alignment based on shared diagonal information is performed, hits are pre-clustered and second the remaining sequences are aligned using Foldseek’s structural Smith-Waterman. (5) The remaining structures meeting alignment criteria are clustered using MMseqs2’s clustering module. After the Linclust step the centroids are successively clustered by three cascaded steps of prefiltering, structural Smith-Waterman alignment and clustering using Foldseek’s search.