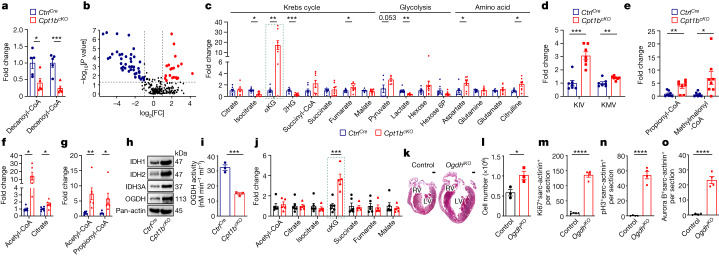
Fig. 3. Impeded mitochondrial import of fatty acid rewires the metabolism of CMs and boosts αKG levels.
a, Metabolic flux assay of CtrlCre and Cpt1bcKO hearts perfused with [13C]palmitate (n = 5 each). b, Volcano plot showing different metabolites in CtrlCre and Cpt1bcKO hearts (n = 8 each). Symbol colors indicate increased (red), decreased (blue) or unchanged (black) metabolites in Cpt1bcKO CMs. Dashed horizontal line indicates the threshold of changed metabolites with a P value of < 0.05; dashed vertical lines indicate a fold change of >2 or <0.5. c, Quantification of metabolites associated with the Krebs cycle and glycolysis, as well as amino acids, in Cpt1bcKO hearts (pyruvate, n = 6 each; all other metabolites, n = 8 each). Dashed outline indicates the change in αKG. d,e, Quantification of BCAA catabolism-associated metabolites (n = 7 for d; n = 8 for e). KIV, α-ketoisovalerate; KMV, α-keto-β-methylvalerate. f,g, Metabolic flux assay of CtrlCre and Cpt1bcKO hearts perfused with [13C]glucose (f) or [13C]isoleucine (g) (n = 6 each). h, Western blot analysis of enzymes involved in αKG generation and catabolism (n = 3 each). Pan-actin was used as a loading control. For gel source data, see Supplementary Information. i, Enzymatic activity of OGDH in CMs from adult CtrlCre and Cpt1bcKO mice (n = 3 each). j, Comparison of Krebs cycle metabolites in CMs from control hearts (n = 6) and OgdhiKO hearts (n = 5). Dashed outline indicates the change in αKG k, Trichrome staining of heart sections from control and OgdhiKO mice, 3–4 weeks after termination of TAM treatment for Ogdh deletion. RV, right ventricle; LV, left ventricle. Scale bars, 500 μm. l, Quantification of CMs in adult control and OgdhiKO hearts (n = 3 each). m–o, Quantification of Ki67+ (m), pH3+ (n), aurora B+ (o) and sarc-actinin+ CMs on heart sections from control and OgdhiKO mice (m, control n = 5; OgdhiKO n = 4; n and o, n = 4 each). Error bar represents mean ± s.e.m. n numbers refer to individual mice. Two-tailed, unpaired Student t-tests were used for statistical analysis in a,c–g,i,j,l–o. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.