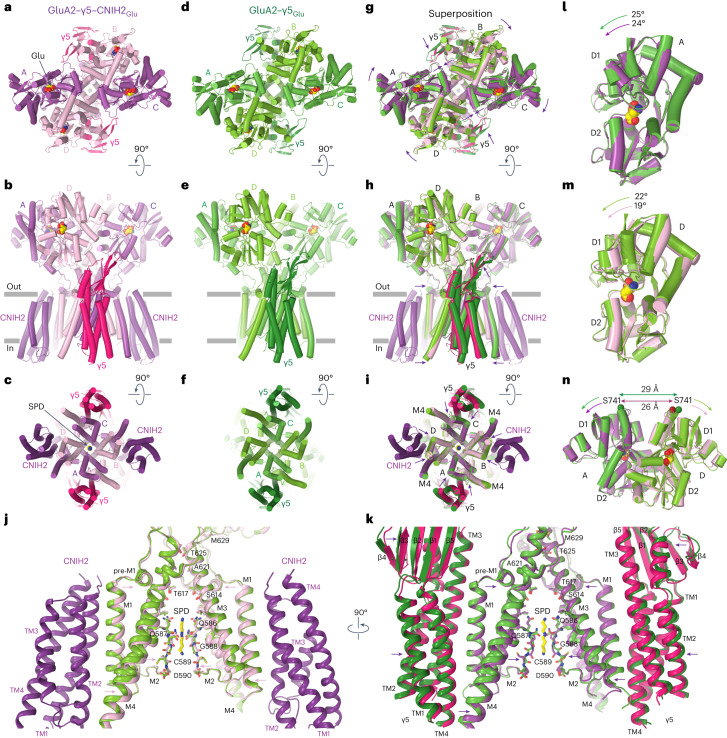
Fig. 8. CNIH2 reduces separation of the upper D1 lobes in LBD dimers during desensitization.
a–i, Structures of GluA2–γ5–CNIH2Glu (a–c), GluA2–γ5Glu (d–f) and their superposition based on the GluA2 TMD (g–i) viewed extracellularly (a,d,g), parallel to the membrane (b,e,h) and intracellularly (c,f,i), with molecules of Glu and SPD shown as space-filling models. Relative movement of domains upon CNIH2 binding is illustrated with purple arrows. j,k, GluA2–γ5–CNIH2Glu and GluA2–γ5Glu TMD superposition viewed parallel to the membrane from two perpendicular directions, with GluA2 subunits A and C and γ5 molecules (j) and GluA2 subunits B and D and CNIH2 molecules (k) omitted for clarity. The pore-lining residues and the molecule of SPD are shown as sticks. Arrows indicate tightening of the TMD in the presence of CNIH2. l,m, D2 lobe-based superposition of subunits A (l) and D (m) LBDs from GluA2–γ5–CNIH2Glu (purple) and GluA2–γ5Glu (green) structures. Arrows show rotation of the upper lobe D1 toward lower lobe D2 relative to the close-state structures GluA2–γ5–CNIH2ZK and GluA2–γ5ZK. n, D2 lobe-based superposition of subunits A/D LBD dimers from GluA2–γ5–CNIH2Glu (purple) and GluA2–γ5Glu (green) structures. Double arrows indicate the distance between Cα atoms of S741.