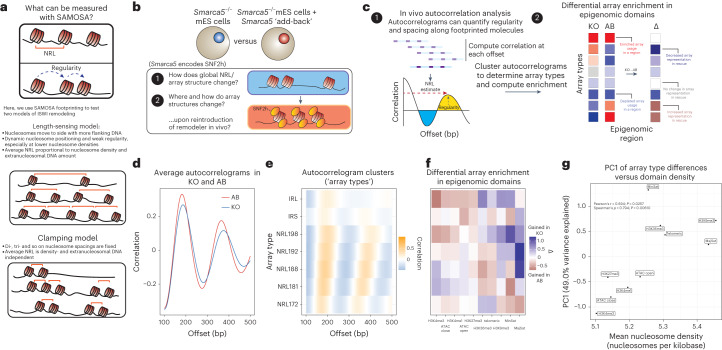
Fig. 1. Measuring structural consequences of SNF2h rescue in mES cells at the resolution of single nucleosome arrays.
a, Schematic overview of the structural features of nucleosome arrays measurable using the SAMOSA approach. We aimed to use SAMOSA to distinguish between two possible models of remodeling by ISWI-family, nucleosome-sliding remodelers. b, Experimental design of our in vivo footprinting experiment, wherein we footprint mES cells devoid of the SNF2h ISWI ATPase subunit (KO cells), and cells where the SNF2h ATPase has been reintroduced through cDNA overexpression (AB cells). We then ask how NRLs on individual fibers change across epigenomic domains. c, Schematic of our analytical pipeline, where we calculate single-molecule autocorrelations (left), which effectively measure the NRL and regularity of individual footprinted molecules, and then perform Leiden clustering and differential enrichment analysis (right) to determine how the reintroduction of SNF2h impacts the distribution of arrays observed across specific epigenomic domains. d, Average single-molecule autocorrelograms for AB (red) and KO (blue) samples. AB cells have an NRL estimate of 182 bp, and KO cells have an NRL estimate of 187 bp. e, Average single-molecule autocorrelograms following Leiden clustering of individual molecules. We observe seven different array types, ranging from NRL172 to NRL198, as well as two irregular array types we term IRL and IRS. f, Differential array enrichment across ten different epigenomic domains; red indicates gained array type usage in AB cells, and blue indicates gained array type usage in KO cells. ATAC close refers to sites that close upon rescue of SNF2h activity; ATAC open refers to sites that open upon rescue of SNF2h activity. g, Following PCA reduction of the matrix in f, we correlated PC1 against the average single-fiber nucleosome density of each domain analyzed in f. PC1 significantly correlates with average nucleosome density of studied domains (two-sided test).