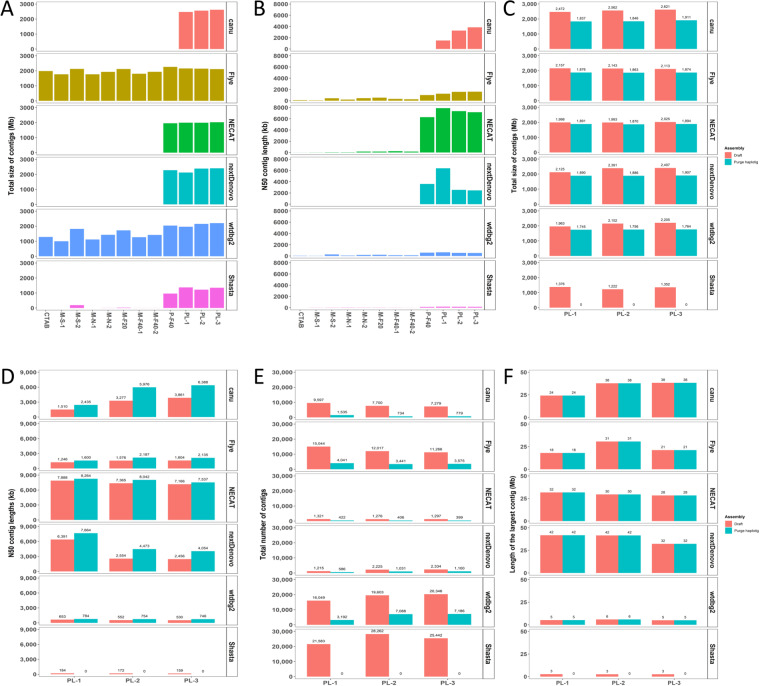
Fig. 2.
Assembly statistics for Nanopore sequencing. (A) Assembled genome size of each genome assembler. The x-axis indicates nanopore sequencing batch and the y-axis indicates total contig size. (B) Contig N50 of assembled genomes. The x-axis indicates nanopore sequencing batch and the y-axis indicates contig N50 length of the genome. (C) Assembled genome size for Draft and Purge Haplotigs-processed genomes. (D) Read N50 for Draft and Purge Haplotigs-processed genomes. (E) Number of contigs in the Draft and Purge Haplotigs-processed genomes. (F) Comparison of the longest contig lengths for Draft and Purge Haplotigs-processed genomes.