Figure 2.
Identified PD modules from RNAseq of skin biopsy
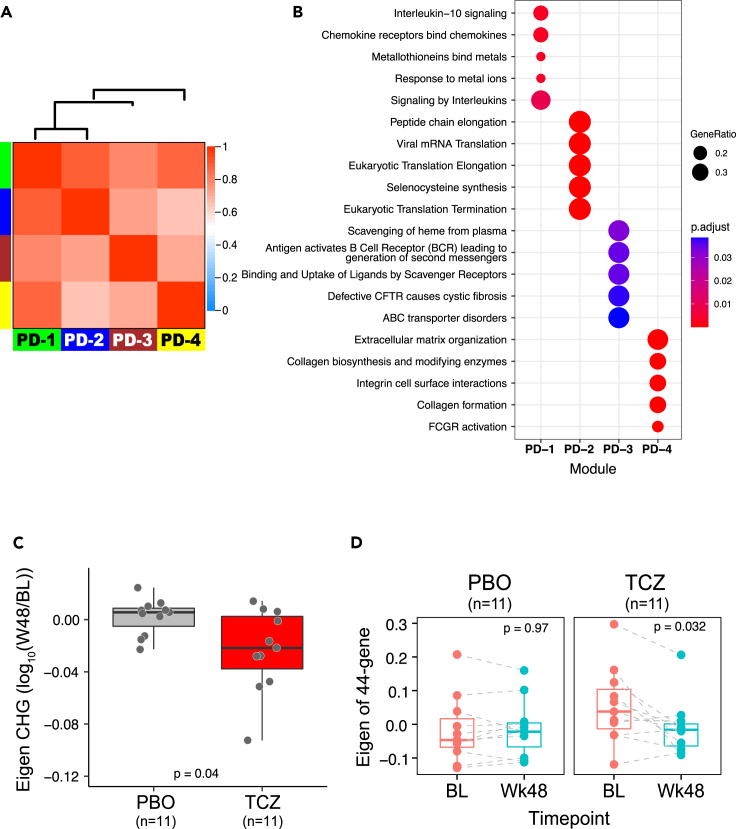
(A) The Module eigengene adjacency dendrogram (top) and heatmap with red and blue indicating highly related and unrelated modules, respectively, depicting the relationship of the modules of PD genes identified by WGCNA. The color row below the heatmap indicates PD module assignment: PD-1 (n = 33 genes), PD-2 (n = 211 genes), PD-3 (n = 44 genes), and PD-4 (n = 44 genes).
(B) Reactome pathway enrichment of each module gene set. Pathways were determined for each module gene using the compareCluster function with Reactome database. The top 5 of the most over-represented the classification of pathways are illustrated as dot plots, with the gene ratio denoted by size and the significance denoted by color. The p values were adjusted by the Benjamini-Hochberg method.
(C) PC module eigengene (ME) was down-regulated in TCZ treatment group compared with placebo group in the FocuSSced cohort. The eigengene change of PC module 48 weeks post treatment from pretreatment is expressed as log10 on y axis; the p value (Wilcoxon test) is indicated at the bottom of graph.
(D) Paired comparison of module eigengene at baseline and 48 weeks following-up. Intra-patient comparison showed a significant reduction of ME in patients with TCZ treatment but not with placebo (PBO).