Figure 5.
Skin PC signature enriched in SSc-ILD lung
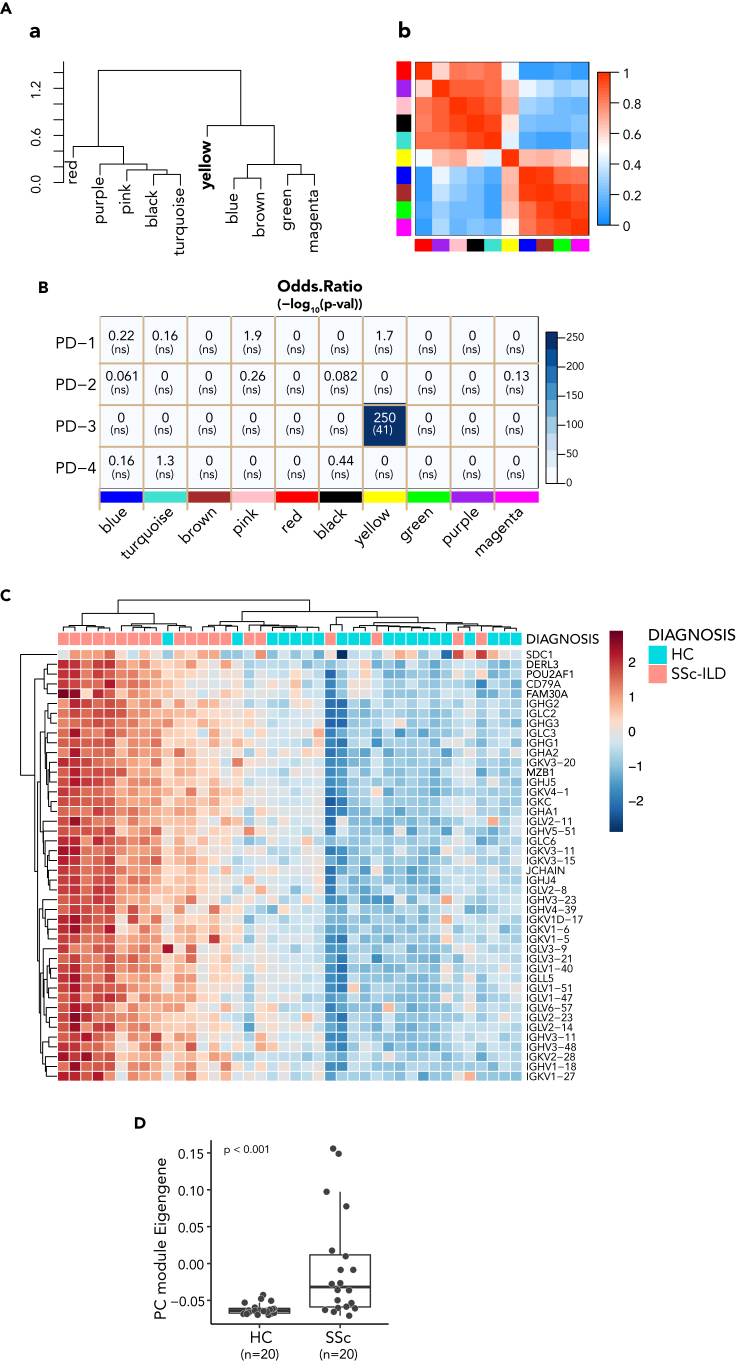
(A) Relationship between WGCNA derived module eigengenes from RNAseq of SSc-ILD lung.
(a) The eigengene dendrogram of DEG genes on hierarchical clustering of adjacency-based dissimilarity, and (b) relationship of modules on eigengene adjacency heatmap. The modules assigned in WGCNA were either text labeled in (a) or color labeled on both axes of the heatmap in (b), respectively. Colors in heatmap are from low adjacency (blue) to high adjacency (red).
(B) Heatmap showing the overall results from overlap analysis between the module genes from SSc-ILD lung and PD modules from SSc skin. Blue color gradient represents the odds ratio score. The number on the top in each square box of heatmap represents the odds ratio value from the analysis, while the number in the parentheses at the bottom represents the p value in -log10. The color labeled modules derived from the RNAseq of SSc-ILD lung are on the x axis and 4 PD modules from skin RNAseq are on y axis. ns represents no statistical significance with a cutoff of p < 0.05.
(C) Heatmap of elevated PC gene expression in RNAseq of SSc-IL lungs compared with control lungs. PC module genes (n = 44) in rows and subjects in columns were hierarchically clustered and the gene expression was transformed and standardized to Z score. The conventional PC marker CD138 (encoded by SDC1) was included as a reference displayed on the top of row in the heatmap. The four PC signature genes are highlighted in bold in the label.
(D) Elevated PC ME in lung of SSc-ILD. PC ME of SSc-ILD lung was compared with controls. p value shown on the top of plot was derived from Wilcoxon test.