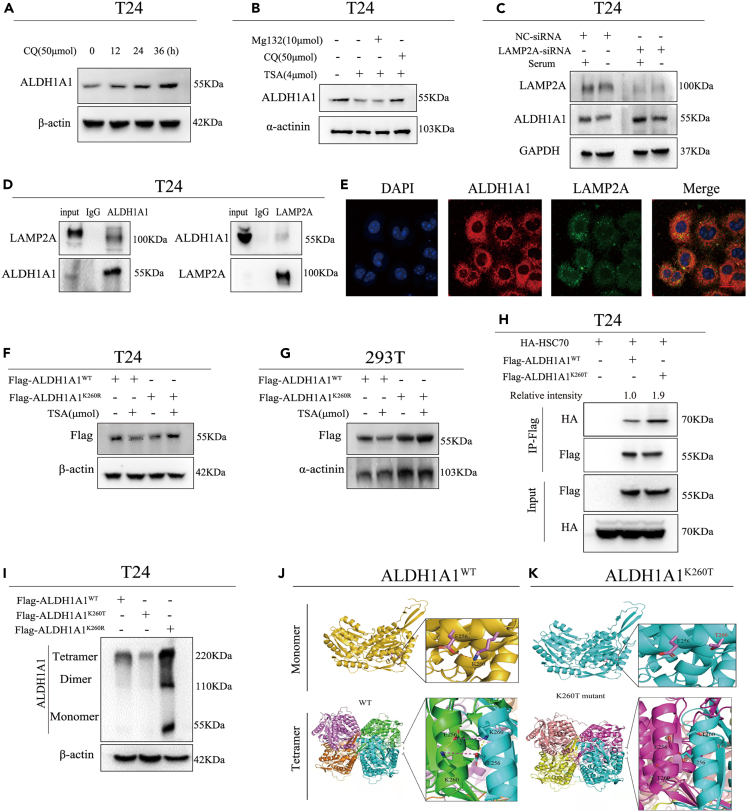
Figure 5.
2-Hydroxyisobutyrylation at lys-260 promoting ALDH1A1 degradation through chaperone-mediated autophagy
(A) T24 cells were treated with CQ, and the ALDH1A1 protein levels were detected by immunoblotting at the indicated time points.
(B) MG132 or CQ was cotreated with TSA in T24 cells to observe the protein levels of ALDH1A1 by Western blotting.
(C) LAMP2A was knocked down in T24 cells by siRNA, and the knockdown efficiency of LAMP2A and ALDH1A1 protein levels were examined by Western blotting.
(D) Endogenous LAMP2A was IPed from T24 cells. Rabbit IgG was used as a control.
(E) Colocalization of ALDH1A1 and LAMP2A in T24 cells was observed by confocal microscopy. Scale bar = 20 μm.
(F) TSA treatment decreased the level of ALDH1A1 wild-type, but not K260R mutant ALDH1A1 in T24 cells.
(G) TSA treatment decreased the level of ALDH1A1 wild-type, but not K260R mutant ALDH1A1 in 293T cells.
(H) HA-tagged HSC70 was co-transfected with vector, flag-tagged ALDH1A1WT, or flag-tagged ALDH1A1K260T into T24cells, the ALDH1A1-HSC70 binding was determined by immunoprecipitation-Western blotting analysis.
(I) The cell lysates were collected with ALDH1A1WT, ALDH1A1K260T, or ALDH1A1K260R overexpressed in T24 cells followed treatment with 1% glutaraldehyde, while the polymerizations of ALDH1A1 were examined by immunoblotting.
(J and K) The crystal structures of ALDH1A1WT and ALDH1A1K260T were analyzed by AlphaFold 2.0 software. For cell experiments, each experiment was performed at least three times.