Figure 3.
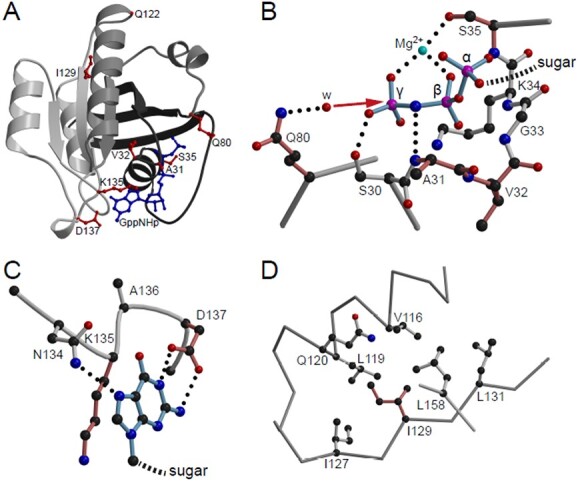
Structural visualization of Rab5c variants. (A) Crystal structure of Rab5c bound to the hydrolysis-resistant GTP analogue GppNHp (pdb-entry 1HUQ). Tints of gray are used to illustrate the incompatibility of the non-sense variants with protein folding; dark gray, N-terminal part till residue 80; light gray C-terminal part from residue 122 onward. GppNHp (blue) and residues affected by point mutations (red) are shown in ball-and-stick representation. (B) Detailed view of the phosphate binding cavity. For clarity, only the three phosphates of the nucleotide are shown and labeled α, β and γ. The direction of the nucleophilic attack of a water molecule (w) to the γ-phosphate in course of the non-GAP catalyzed hydrolysis reaction is indicated by a red arrow. Residues affected by point mutations are colored in a tint of red. Dotted lines, hydrogen bonds. (C) Interaction of the base with the NKxD motif. For clarity of representation, only the base of the nucleotide is shown. (D) Hydrophobic environment of Ile129. Figures were generated by use of the programs molscript (55) and Raster3D (56).
