Figure 6 – The TET1/TDG-related inflammatory signature separates human colon adenocarcinoma cases according to their genomic instability features; concerted high DNMT1 expression and low TET1 expression in human CIMP-H colonic adenocarcinoma.
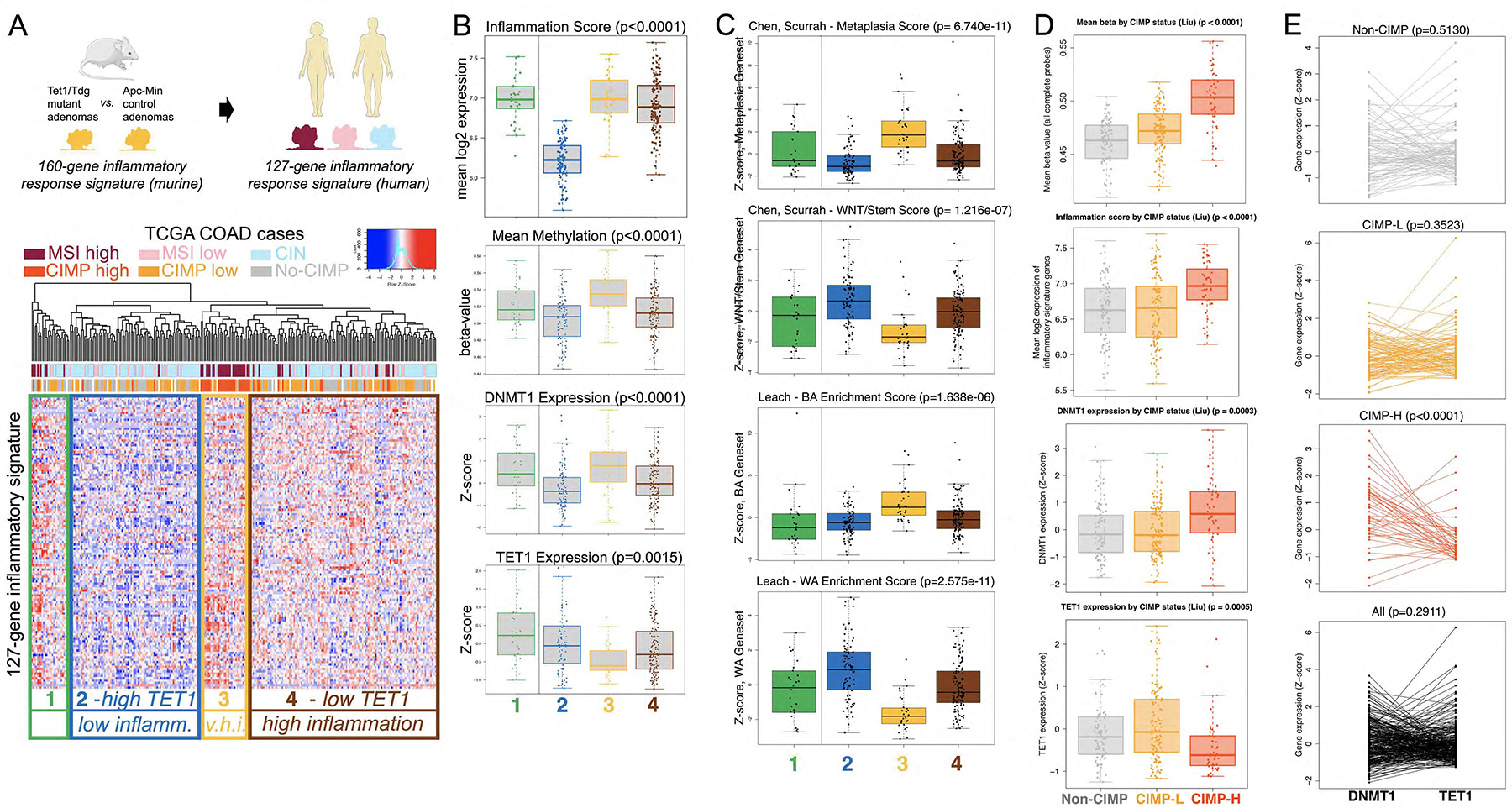
(A) The 127-gene human TET1/TDG-related inflammatory signature, derived from the 160-gene murine Tet1/Tdg-inflammatory signature, identifies 4 clusters in TCGA COAD (colon adenocarcinoma) differing in microsatellite instability (MSI) or chromosomal instability (CIN) status. V.h.i.=very high inflammation. (B) Box plot representation of inflammation score (expression levels of 127-gene signature), mean methylation, DNMT1 and TET1 expression levels in the 4 clusters. (C) Box plot representation of expression (Z-score) of Metaplasia and WNT/Stem signatures from Chen, Scurrah et al.48, and BA (Braf-Alk5) and WA (Wnt activation) signatures from Leach et al.49. (D) Box plot representation of mean methylation (β-value), inflammation score (expression levels of 127-gene signature), DNMT1 and TET1 expression levels in TCGA COAD cases, separated according to CpG Island Methylator Phenotype (no-CIMP, CIMP-L, and CIMP-H, in gray, orange and red, respectively, as in panel A). (E) Relationship between DNMT1 and TET1 expression levels in TCGA COAD cases (mRNA expression Z-scores from cBioPortal; from top to bottom: no-CIMP, CIMP-L, CIMP-H and all cases); DNMT1 expression was higher than TET1 expression in CIMP-H samples (36/46 cases, 78.3%, p-value< 0.0001 for paired Wilcoxon test).
