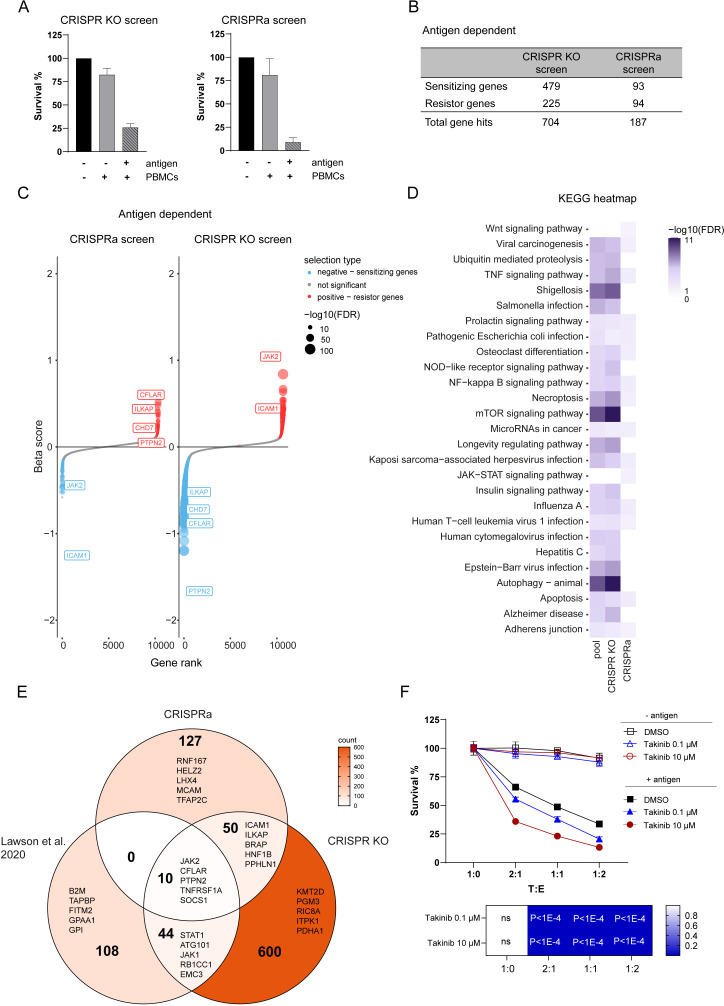
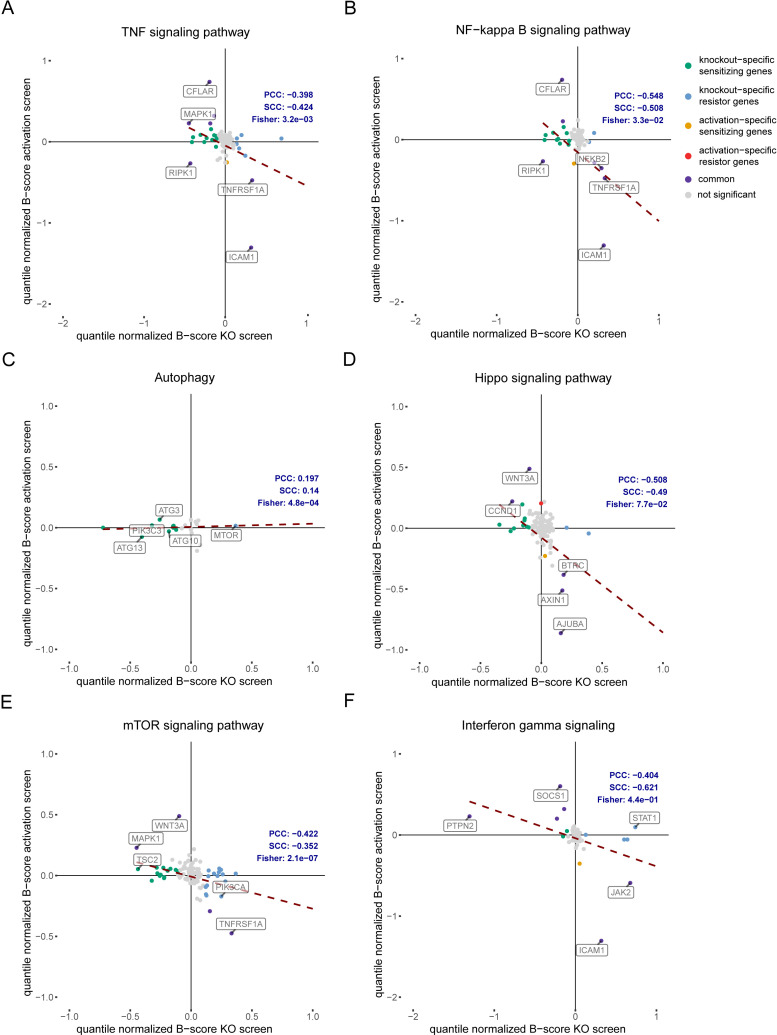
Figure 3. Discovery of genes regulating tumor resistance and sensitivity to CTL killing.
(A) Cell survival after co-culturing with PBMCs containing antigen specific CTLs for 3 days normalized to tumor cells not exposed to PBMCs and antigen for CRISPR KO (left) and CRISPRa (right) screen. (B) Table displaying the numbers of gene hits specific for antigen-dependent setup identified by CRISPR KO and CRISPRa screen. (C) Ranked-ordered, beta-scores for antigen-dependent screen setup (CRISPRa – left; CRISPR KO – right). The top best scoring overlapping gene hits between CRISPR KO and CRISPRa screen are indicated. Hits at FDR <5% are highlighted in red (positive selection - resistor genes) and blue (negative selection - sensitizing genes). (D) KEGG pathway enrichments for top 15 best scoring pathways in CRISPR KO, CRISPRa or pooled screen hits represented as heatmap: white – not statically significant (FDR corrected hypergeometric overrepresentation test). (E) Venn diagram displaying intersection of CRISPRa screen gene hits, CRISPR KO screen gene hits and previously published tumor resistance core gene data set of Lawson et al., 2020. Top 5 ranked genes were indicated in each sector. (F) Tumor killing assay in the presence of different concentrations of TAK1 inhibitor (Takinib) as indicated or DMSO control and cell survival was measured after 3 days (top). Bar graphs show normalized mean ± SD in triplicate representative for two independent experiments. Two-way ANOVA corrected for multiple comparison according to Dunnett was used to determine statistical significance (bottom) (ns: not significant).