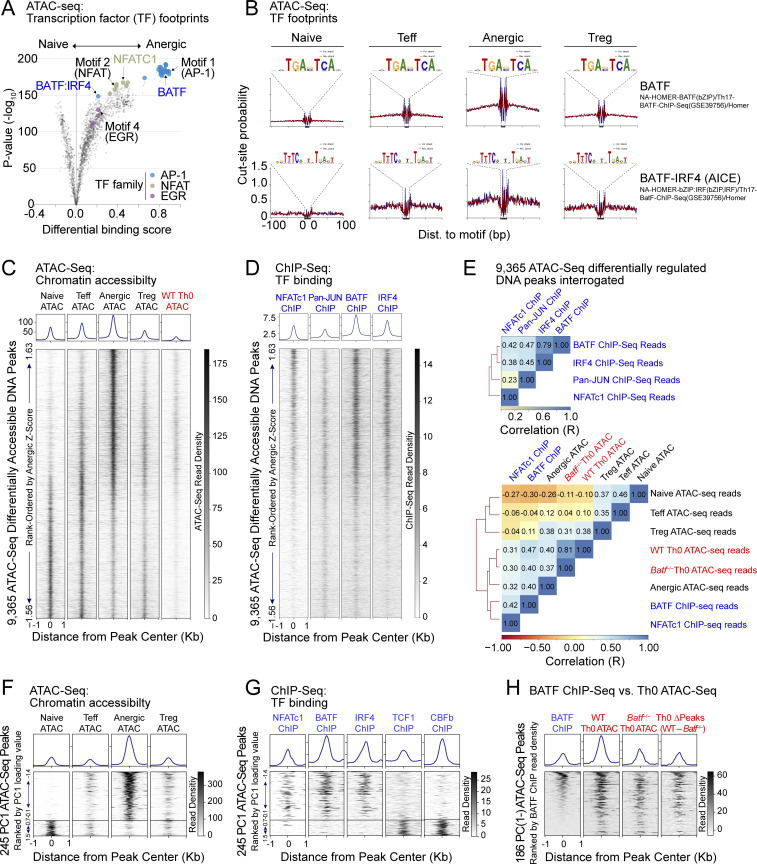
Figure 2.
BATF and IRF4 co-localize at BATF-IRF4 (AICE) motifs in anergic T cells. (A) Volcano plot summary of TF footprint enrichments determined by TOBIAS (Bentsen et al., 2020) from pairwise comparison of CD44low Foxp3(GFP)− naïve and CD44high Foxp3(GFP)− CD73high FR4high Nrp1+ anergic samples across 38,509 consensus ATAC-seq peaks. Colors denote footprints corresponding to anergy-associated TF families (as indicated), including the de novo motifs identified in Fig. 1. (B) Visualized footprints for BATF (top) and the BATF-IRF4 AICE (bottom) based on the ATACseqQC algorithm. Plots display Tn5 transposase cut-site probability within ±100 bp of all consensus binding motifs identified on chromosome 1 for both forward (blue) and reverse (red) de-duplicated and shifted ATAC-seq reads in each sample group (naïve, Teff, anergic, and Treg). (C and D) ATAC-seq read densities (C) and ChIP-seq read densities (D) within ±1 kb of the peak center for 9,365 differentially-accessible chromatin regions, displayed as a stack according to anergic Z-score. Histograms above represent the average read density across the same intervals for each sample group. ChIP-seq information was derived from public datasets for CD8+ T cell NFATc1 (Klein-Hessling et al., 2017), CD4+ T cell Pan-JUN (Li et al., 2012), CD4+ T cell BATF, and IRF4 (Iwata et al., 2017). 72 h in vitro stimulated (anti-CD3/CD28 mAb) WT Th0 ATAC-seq reads were mapped from publicly available ATAC-seq SRA files (Karwacz et al., 2017). (E) Pairwise Spearman correlation (R) and hierarchical clustering of ChIP-seq reads (top) and similar analysis of ATAC-seq reads as compared to BATF or NFATc1 ChIP-seq (bottom), when interrogated at 9,365 differentially accessible ATAC-seq peak intervals as shown in C and D. Batf−/− Th0 ATAC-seq reads were previously reported (Karwacz et al., 2017). (F) A total of 245 unique ATAC-seq peaks assigned to PC cluster groups PC(1−) and PC(1+) after TSNE plotting were ranked according to lowest PC1 loading value, and then were displayed as a stack with read densities in each sample group as indicated (lower panels). Histograms sum all reads across all selected peak intervals (upper panels). (G) Published ChIP-seq datasets for NFATc1, BATF, IRF4, TCF1, and CBFb were interrogated across the identical PC(1−) and PC(1+) ATAC-seq peak coordinate intervals as shown in F, with ChIP-seq read densities again displayed as stacks. (H) Published BATF ChIP-seq reads at 186 unique PC(1−) ATAC-seq peaks were ranked by highest BATF read density and then compared to published WT and Batf−/− Th0 ATAC-seq reads following 72 h of in vitro stimulation with anti-CD3/CD28 mAbs. One additional stack (Th0 ΔPeaks) represents the numerical difference between the WT and Batf−/− ATAC-seq read counts at each DNA coordinate. Source ATAC-seq data for all panels as in Fig. 1.