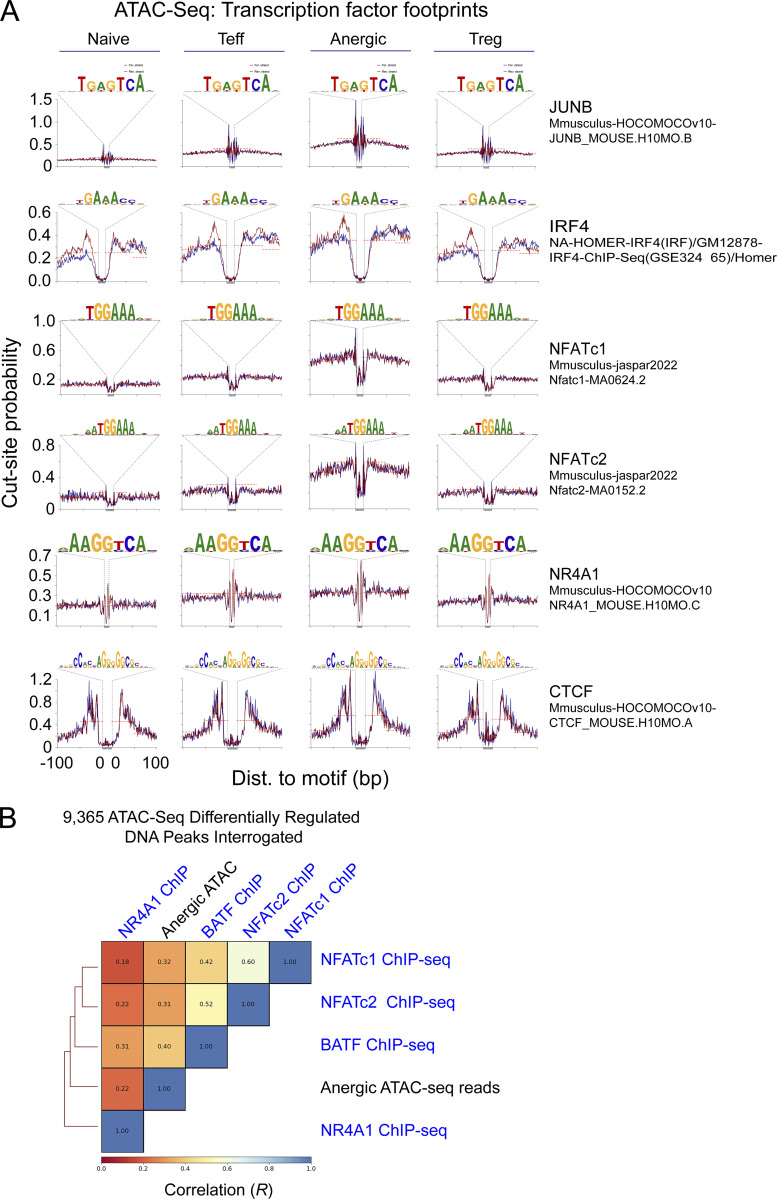
Figure S1.
ATAC-seq chromatin accessibility peaks are bound by BATF, IRF4, and NFAT proteins in anergic CD4+ T cells. (A) Using the ATACseqQC algorithm, chromosome 1 ATAC-seq reads were interrogated for the presence of JUNB, IRF4, NFATc1, NFATc2, NR4A1, and CTCF consensus DNA-binding motifs. Tn5 transposase cut-site probability across these sequences was then determined for both forward (blue) and reverse (red) de-duplicated and shifted DNA reads in each sample group (naïve, Teff, anergic, and Treg) as indicated. Cut-site protection (“footprint”) when present is indicated by the dashed line. (B) Pairwise Spearman correlation (R) and hierarchical clustering of anergic ATAC-seq reads and select ChIP-seq reads interrogated at 9,365 differentially accessible ATAC-seq peak intervals. Source ATAC-seq data for all panels as in Fig. 1.