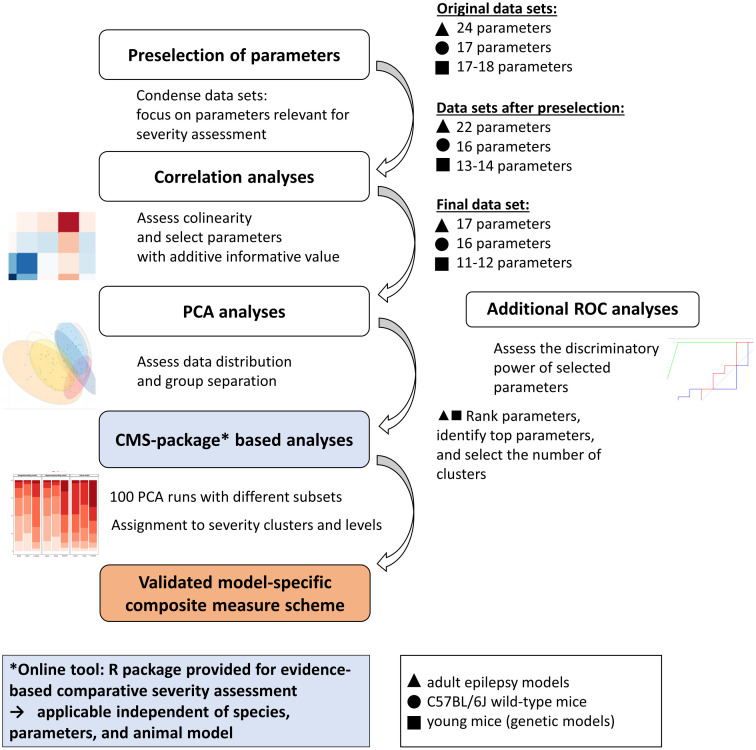
Fig 1. Overview of the bioinformatics workflow.
Combined data sets are subjected to a bioinformatics workflow comprising a preselection of parameters, correlation, and principal component analyses. The basic principles of the step-by-step bioinformatics workflow are illustrated on the left side. The right side of the graph describes their practical application to data sets considered in this publication. The approach allowed a reduction of parameters, which contributed to condensation of information on a distress-related model- and laboratory-specific severity. Further analyses, as displayed in the blue boxes, were computed using a newly designed R package: composite-measure-scheme-(CMS)-package, available at: https://github.com/mytalbot/cms. CMS-package-based analyses allow the ranking and selection of parameters, definition of clusters, and the assignment of individual animal data to model-specific severity clusters. The approach can be applied for a comparison of different models and the assessment of the impact of potential refinement measures. In this context, it should always be noted that laboratory-specific factors can influence severity clustering for a given model. In addition, model-specific receiver operating curve (ROC) analyses allow testing for adequate discriminatory power. This study applied ROC analyses to data sets with three selected behavioral paradigms (Irwin test, open field test, sweetness preference).