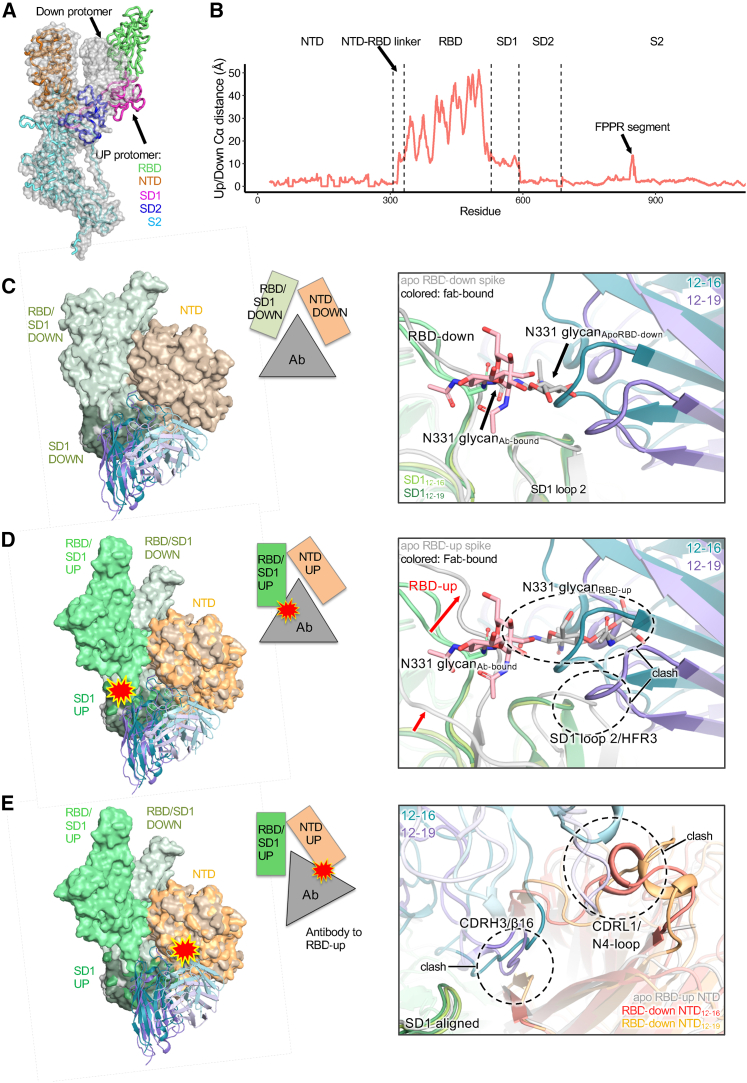
Figure 3.
Antibody binding is incompatible with RBD-up state, suggesting that the antibody is “locked” in a down state
(A) Overlay of an RBD-down (surface) and RBD-up (ribbons) SARS-CoV-2 spike structure (PDB: 7KRR).
(B) The Cα distance for each residue in the spike between the RBD-up and RBD-down state.
(C) The 12-16 and 12-19 Fab structures (colored) were superimposed onto an apo SARS-CoV-2 spike structure (PDB: 6XM5) with RBD-down (gray). When the RBD is down and the Fab is bound, the N331 glycan moves out of the way and is nicely accommodated next to the antibody heavy-chain FR3 region.
(D) The 12-16 and 12-19 Fab structures (colored) were superimposed onto an apo SARS-CoV-2 spike structure (PDB: 6XM3) with RBD-up (gray). When the RBD moves up, there are clashes between the N331 glycan and the heavy chain and the RBD strand leading to SD1 (residue ∼ 530) and heavy-chain FR3.
(E) The 12-16 and 12-19 Fab structures (colored) were superimposed onto an apo SARS-CoV-2 spike structure with RBD-up (gray) using an alignment of the SD1 region of the epitope (residues 531–588) to simulate an “RBD-up bound antibody.” The RBD-up bound antibody would clash with the original position of NTD (C). In this case, the CDRH3 would clash with the β16 sheet of NTD, and the CDRL1 would clash with the N4 loop. Compared with the position of the NTD in the apo RBD-up structure, the antibody CDRH3 would clash with the β16 sheet of NTD, but it is unclear if the N4 loop would clash.