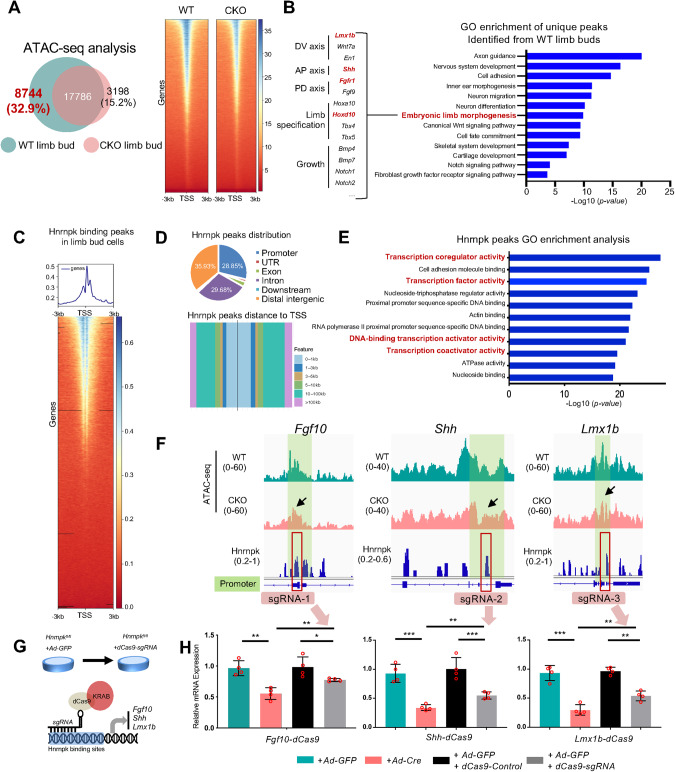
Fig. 4. Hnrnpk maintains the regulation of regulatory axes in a transcriptional manner.
A The number of peaks detected at E11.5 in WT and CKO limb buds by ATAC-seq, and signal intensities of peaks near transcriptional start sites (TSS). N = 3 biological replicates, replicates were merged. B GO enrichment analysis of genes upon the unique ATAC-seq peaks near TSS identified from WT limb buds compared to CKO. The items highlighted were the items we focus on. The genes involved in embryonic limb morphogenesis were shown on left. C Profile and heatmap of Hnrnpk CUT&RUN-seq performed with E11.5 WT limb buds. D The distribution and distances to the TSS of Hnrnpk binding peaks in the WT limb buds. E GO enrichment analysis of the Hnrnpk binding peaks in the WT limb bud. The items highlighted were the items we focus on. F ATAC-seq peaks and Hnrnpk binding at the TSS of Fgf10- (left panel), Shh- (middle panel), and Lmx1b- (right panel) loci in WT and CKO limb buds. The green lines indicated the promoter regions. The red boxes indicated the target of the designed sgRNA. G Schematic diagram of constructing CRISPR/dCas9-KRAB vector. H qPCR results of the Fgf10 (left panel), Shh (middle panel), and Lmx1b (right panel) in groups transfected Hnrnpkfl/fl limb bud cells with dCas9-control or dCas9-sgRNA compared to Hnrnpkfl/fl limb bud cells infected with Ad-GFP or Ad-Cre. N = 4 biological replicates. The p value was calculated by one-way ANOVA with Tukey’s post-hoc test. Data were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.