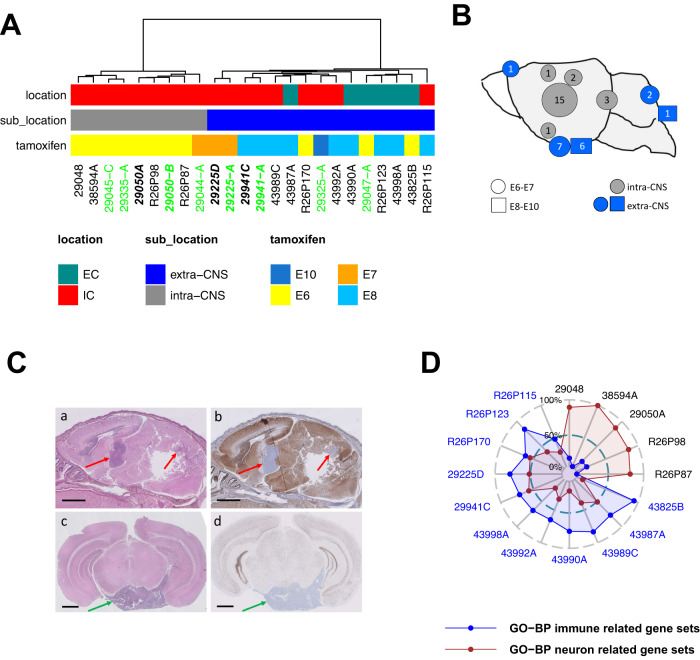
Fig. 3. Genetically engineered mouse models reveal specific anatomical origins for murine Myc and Shh ATRT.
A Unsupervised hierarchical clustering of mouse RT (n = 21 biologically independent mice) based on transcriptomic dataset of combined RNAseq dataset (n = 16 biologically independent samples, sample name colored in black) and gene expression microarray dataset (n = 8 biologically independent samples, sample name colored in green). 3 samples (bold sample names) were processed using the two platforms and used to control the cross-platform data integration. B Anatomical distribution of Rosa26::CreERT2/Smarcb1flox/flox (R26) RT; circles correspond to Tamoxifen injection at E6-E7 while squares correspond to E8-E10; gray indicates intra-CNS tumors, blue, indicates extra-CNS tumors. C Hematoxylin Eosin Safran (HES, a, c) and BAF47 staining (b, d) of sagital (a, b) and coronal (c, d) sections of brains from the R26 mice. Red arrows point to intra-CNS tumors (a, b); green arrows point to extra-CNS tumors (c, d). Scale bars: 1 mm (D) Single sample gene set enrichment analysis of murine RT samples based on RNAseq data (n = 16). Each dot corresponds to one sample; sample names are colored according to molecular subgroups (black: Shh; blue: Myc). Average enrichment scores (in percentage) of the top 5 differentially enriched neuronal development-related (red) and immune-related (blue) gene sets are considered and indicated for each sample. Source data are provided as a Source Data file.