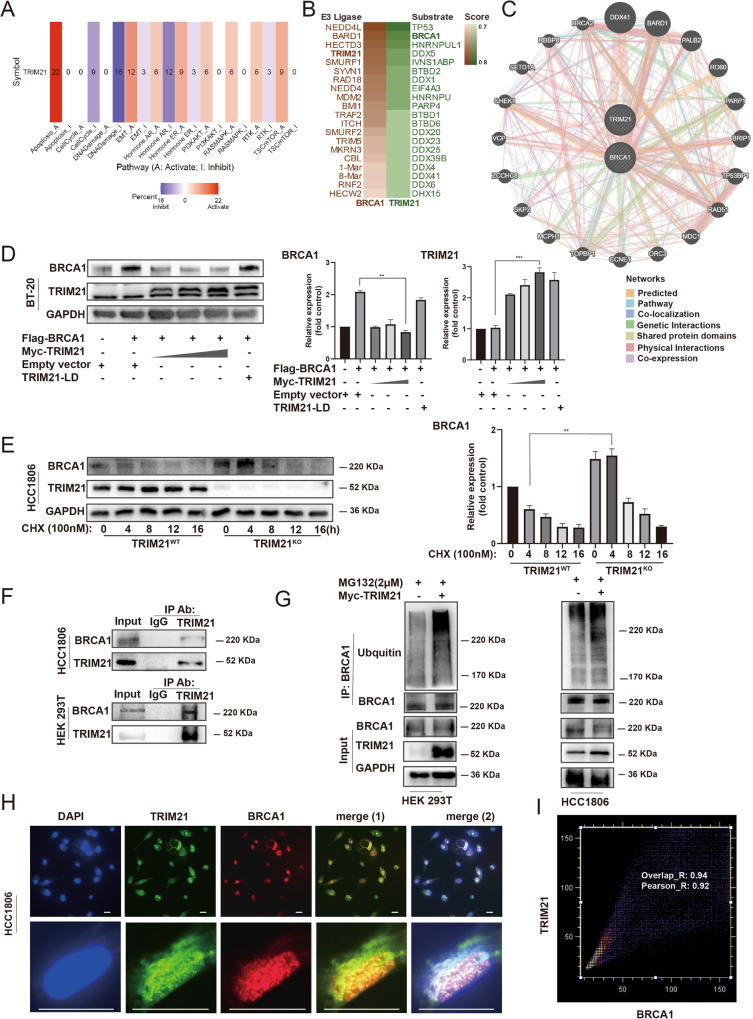
Fig. 2. TRIM21 mediated the ubiquitination degradation of BRCA1.
A A pan-cancer analysis between TRIM21 and signaling pathways by Gene Set Cancer Analysis. B Potential E3 ligases or substrates are predicted by using UbiBrowser. C TRIM21-BRCA1 Interaction analysis was performed in the GeneMANIA database. The specific interaction and predictive values are visible on (http://genemania.org/search/homo-sapiens/TRIM21/BRCA1). D The protein level was examined after transfecting with exogenous full-length BRCA1, TRIM21, and Ligase-dead TRIM21. The GAPDH expression is the loading control. E BRCA1 protein degradation rate in TRIM21WT and TRIM21KO cells. Data were shown as mean ± SD. The significant level was identified by *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. One-way ANOVA analysis is used for (D) and (E). F The CO-IP results between TRIM21 and BRCA1. G 293T cells were transfected with Myc-TRIM21, and cell lysates were subjected to analysis by IP, followed by IB detection. H Immunofluorescent staining revealed the colocalization of TRIM21 and BRCA1 proteins in MDA-MB-231 cells. BRCA1 (red) and TRIM21 (green) and nuclear (blue). Bar length: 10 μm. I Colocalization analysis for immunofluorescent stained nuclear by ImageJ with colocalization plugin JACoP. X-axis: red pixel intensity (BRCA1). Y-axis: green pixel intensity (TRIM21). Overlap_R = 0.94, Pearson_R = 0.92 (calculations were based on data from three independent experiments, at least 15 nucleoli were analyzed).