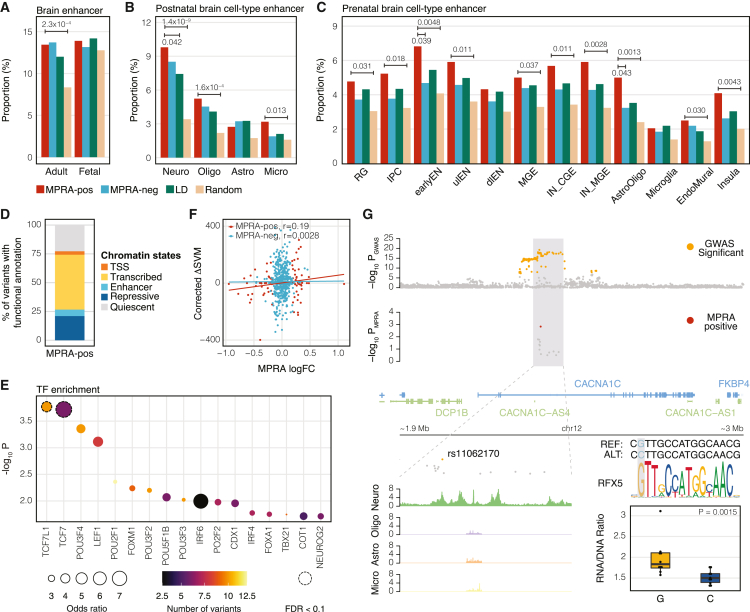
Figure 2.
Epigenetic characterization of MPRA-positive schizophrenia risk variants
(A–C) The proportion of epigenetic overlap of MPRA-positive (MPRA-pos) and MPRA-negative (MPRA-neg) variants, LD SNPs, and random SNPs to the adult and fetal brain enhancers (A), cell-type-specific enhancers in the adult brain (B), and cell-type-specific enhancers in the fetal brain (C). p values were calculated by one-sided Fisher’s exact test. Comparisons have been made between MPRA-positive variants and other sets of variants. Only significant p values are depicted. Neuro, neurons; Oligo, oligodendrocytes; Astro, astrocytes; Micro, microglia; RG, radial glia; IPC, intermediate progenitor cells; earlyEN, early excitatory neurons; ulEN, upper-layer excitatory neurons; dlEN, deep-layer excitatory neurons; MGE, medial ganglionic eminence; CGE, caudal ganglionic eminence; IN, inhibitory neurons.
(D) Proportion of MPRA-positive variants annotated by 15-core chromatin states.
(E) TFs whose motifs are predicted to be altered by MPRA-positive variants. TF enrichment was calculated by comparing TF binding motifs between MPRA-positive variants and LD SNPs. Each dot is color-coded based on the number of variants that are predicted to alter TF binding motifs, and the size of the dot represents the odds ratio. Dotted circles represent TFs that meet the FDR threshold (FDR < 0.1).
(F) Expression outcome of MPRA (measured by MPRA logFC) can be predicted by the combination of TF binding, activity, and expression (measured by corrected ΔSVM) for MPRA-positive variants, but not for MPRA-negative variants. r stands for Pearson’s correlation coefficient.
(G) Integration of MPRA and other functional genomic datasets unveils a causal variant (rs11062170), a trans regulator (RFX5), and a cell type (neuron) for the CACNA1C locus. The alternative allele (C) of rs11062170 breaks the binding motif of RFX5 and is correlated with lower expression of a reporter gene in MPRA.