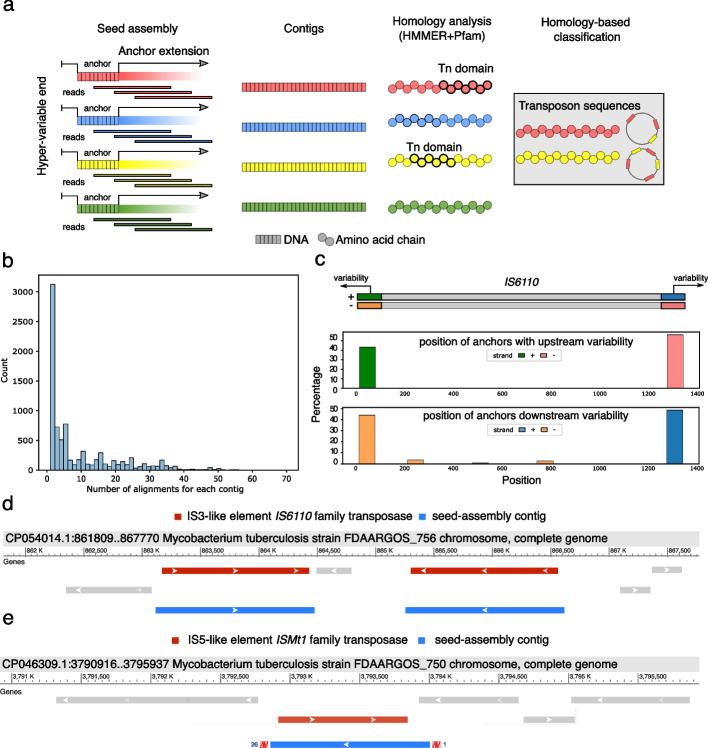
Fig. 3.
De novo discovery of transposons. a DIVE anchors with variability in a single direction are chosen as putative transposon termini that can be used as seeds in an seeded assembly step. The resulting contigs are translated in silico to produce amino acid sequences. Lastly, protein domain homology can be performed on the resulting protein sequences to identify putative transposons, as well as other elements of interest. b Histogram of the number of alignment loci for each contig in at least a M. tuberculosis genome in the NCBI nucleotide database. c Pile up of anchors in IS6110 concentrating on the ends of the insertion sequence, which has a length of 1354 bp. d BLAST alignment of contig104 against a M. tuberculosis genome, mapping to two copies of the element inserted in opposite directions. This contig corresponds to the insertion sequence IS6110. e BLAST alignment of contig2 against a M. tuberculosis genome. This contig corresponds to the insertion sequence ISMt1