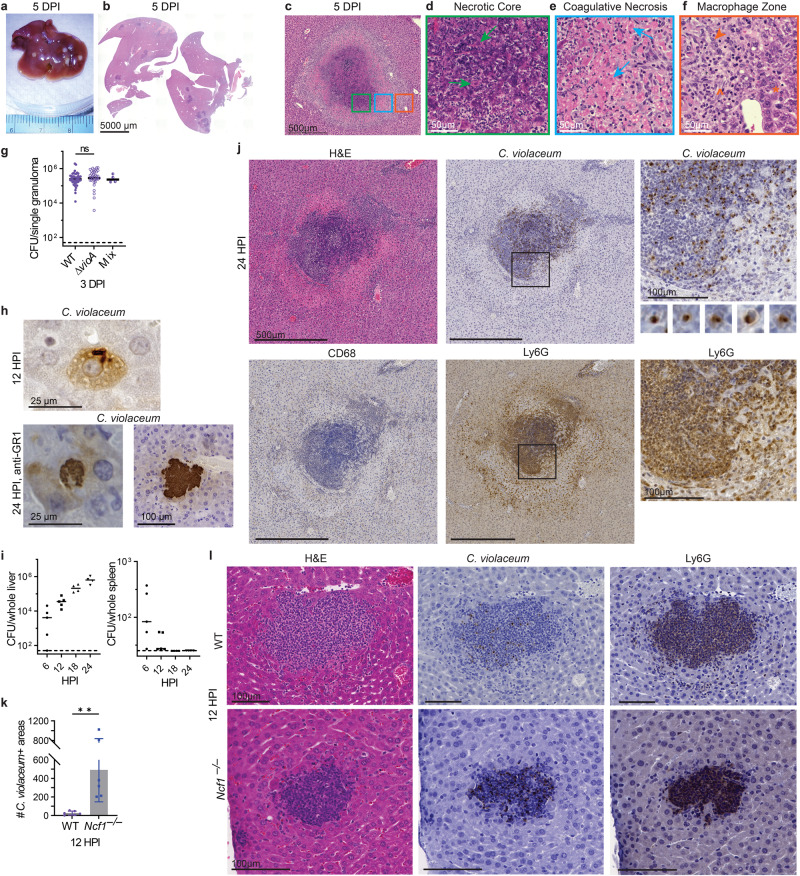
Fig. 1. C. violaceum replicates despite a neutrophil swarm.
a–l Mice were infected with 104 CFU WT C. violaceum, strains indicated. Dashed line, limit of detection; solid line, median. a–f Gross pathology and H&E staining of WT livers 5 dpi. Representative of 10 experiments, each with 3–4 mice, each with multiple granulomas per section. Arrows, coagulative necrotic hepatocyte; arrowhead, macrophage; carrot, fibroblast; asterisk, healthy hepatocytes. g Inoculum was a 1:1 mixture of WT and ∆vioA C. violaceum. 83 individual granulomas were dissected 3 dpi, bacterial burdens determined, and scored for color. Data combined from two experiments. N.s., not significant by Kruskal–Wallis test. h Visualization of C. violaceum within a hepatocyte at 12 hpi by IHC and within hepatocytes in anti-GR1-treated mice at 24 hpi by IHC. For anti-GR1 depletion infections, mice were treated with either anti-GR1 or isotype control at day −1 and 0 and harvested at 24 hpi, with 4 mice per treatment group. Data representative of two experiments. i Bacterial burdens in the liver and spleen at indicated timepoints. Representative of 2 experiments; each point is a single mouse. j Serial sections of WT liver stained by H&E or indicated IHC markers 1 dpi. Representative of 5 experiments, each with 4 mice, each with multiple granulomas per section. k Quantitation of C. violaceum-positive stained areas per section 12 hpi in WT and Ncf1–/– mice. Data combined from two experiments, each with three mice per genotype. Bar represents mean ± SD. **p = 0.0078, by unpaired two-tailed t test. l Serial sections of WT or Ncf1–/– livers stained by H&E or indicated IHC markers 12 dpi. Representative of 2 experiments, each with 3–4 mice per genotype.