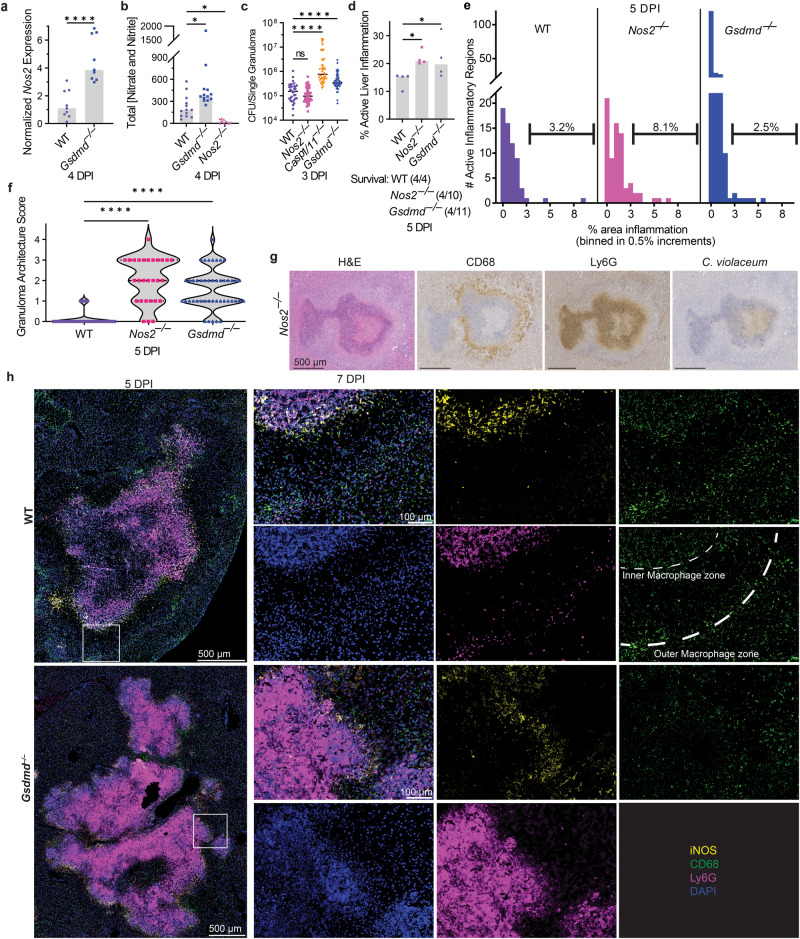
Fig. 7. Gasdermin D and iNOS are independently required to maintain the granuloma architecture.
a–h Mice were infected with 104 CFU WT C. violaceum. a qPCR of Nos2 gene expression in WT and Gsdmd–/– granulomas 4 dpi. Representative of two experiments; each point is 5–6 pooled granulomas harvested from single mouse. ****p < 0.0001, by Mann–Whitney test. b Total nitrate and nitrite concentrations in serum from WT, Gsdmd–/–, and Nos2–/– mice 4 dpi. Representative of two experiments performed in triplicate. *p = 0.0471 (WT vs Gsdmd–/–) or p = 0.0167 (WT vs Nos2–/–), by Kruskal–Wallis test. c Bacterial burdens of single granulomas 3 dpi. Data combined from two experiments; each point is a single granuloma. Solid line, median. n.s. nonsignificant, ****p < 0.0001, by One-way ANOVA test. d, e Percent active inflammation quantified per whole liver section stained with H&E 5 dpi. Representative of two experiments, each with four mice per genotype. d Total percent active liver inflammation; each point is a single mouse. *p = 0.0482 (WT vs Gsdmd–/–) or p = 0.0285 (WT vs Nos2–/–), by Kruskal–Wallis test. Text below the graph denotes mice that died prior to harvest; these mice were not included in the histology analysis. e Percent active liver inflammation per each individual inflammatory region from d. Data combined from all mice within genotype from d. Frequency distribution histogram by the size of inflammatory region, binned in 0.5% increments. Brackets indicate total percentage of inflammatory regions that are above 3% area in size. f Granulomas scored for failure of granuloma architecture 5 dpi. Zero indicates fully intact granuloma architecture; intermediate score includes granulomas with discreet breaches in the macrophage zone; four represents a complete loss of architecture. Data combined from 2 experiments, each with 3, 4 mice per genotype, each with multiple granulomas per section. ****p < 0.0001, by Kruskal–Wallis test. g Serial sections of Nos2–/– liver stained by H&E or indicated IHC markers showing a failed granuloma 7 dpi. Representative of 2 experiments, each 3, 4 mice, each with multiple granulomas per section. h WT and Gsdmd–/– liver sections stained with indicated IF markers 5 dpi. Representative of 2 experiments, each with 4 mice per genotype, each with multiple granulomas per section.