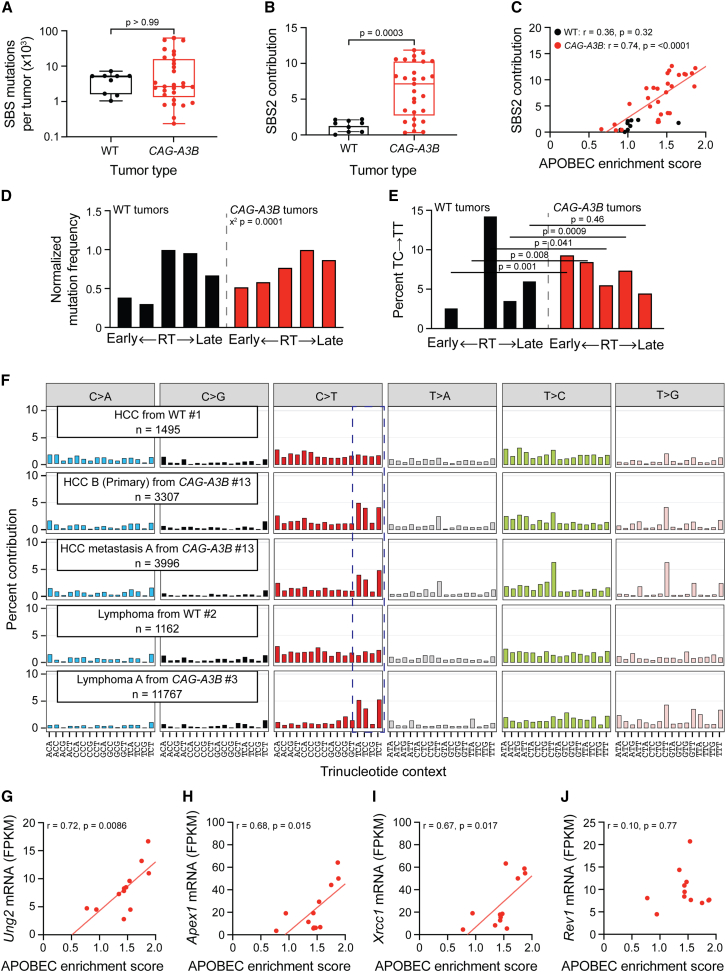
Figure 5.
CAG-A3B tumors exhibit APOBEC3 signature mutations
(A and B) Box and whisker plots of the total number of SBS mutations and the percentage of SBS2, respectively, in tumors from WT and CAG-A3B mice. The middle horizontal line is the median, the top and bottom of the box specify the upper and lower quartiles, and the whiskers outside the box represent the maximum and minimum values (Mann-Whitney U test p value indicated).
(C) Scatterplots comparing APOBEC mutation signature enrichment scores to the percentage contribution of SBS2 in tumors from WT and CAG-A3B animals (Pearson correlation coefficient and p values indicated). Linear regression shown for CAG-A3B data (not possible for WT).
(D) Bar plots showing the proportion of mutations in WT and CAG-A3B tumors according to early- to late-replicating regions (mutation numbers normalized to the largest quintile in each group). The chi-squared test p value is indicated.
(E) Bar plots showing the percentage of TC-to-TT mutations as a percentage of all mutations in each quintile in (D) (Mann-Whitney U-test p values indicated).
(F) Representative SBS mutation profiles for the indicated tumors from WT or CAG-A3B animals (mutation numbers shown). The dashed box highlights APOBEC3-preferred TC motifs characteristic of SBS2.
(G–J) Scatterplots of APOBEC enrichment scores from CAG-A3B lymphomas (n = 12) compared to the mRNA levels of Ung2, Apex1, Xrcc1, and Rev1, respectively, from the same tumors (linear regression lines and Pearson correlation coefficients and corresponding p values indicated).
See also Figures S5–S7 and Table S1.