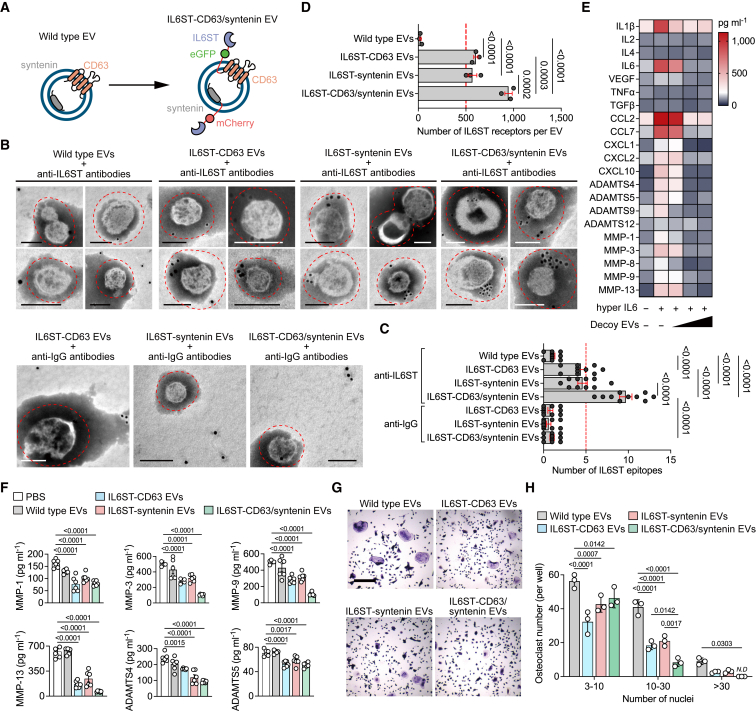
Figure 4.
Co-expression of chimeric proteins optimizes the display of decoy receptors
(A) Schematic diagram of the arrangement at the EV membrane of IL6ST fused to the sorting proteins in tandem with fluorescent proteins.
(B) Representative transmission electron micrographs of decoy EVs with nanogold-labeled antibodies staining of IL6ST. Scale bars, 100 nm. Nanogold-labeled IgG antibodies was used as a negative control. These micrographs are representative of over 10 such images for each group.
(C) Quantification of nanogold-labeled IL6ST epitopes at the EV membrane. The dot plot represents the number of nanogold particles from individual micrographs. n = 10 images each group.
(D) Total estimated number of IL6ST receptors per EV estimated based on obtained MFI values after fluorescence calibration of with Quantibrite PE beads by flow cytometry. n = 3 biological replicates.
(E) Heatmap showing the expression of interleukins, chemokines, and matrix enzymes in the supernatant of mouse cartilage explants treated with 50 ng/mL hyper IL6 and increasing doses of decoy EVs. Each cell of heatmap represents the average expression value of cytokines from three biological replicates. Cytokines were measured using Luminex assay.
(F) Quantitative analysis showing the expression of six matrix enzymes in the supernatant of mouse cartilage explants treated with 50 ng/mL hyper IL6 and 3 × 109/mL decoy EVs. Each dot represents the average expression value of cytokines from three biological replicates. Cytokines were measured using Luminex assay.
(G) Representative TRAP stain images of BMMs treated with 50 ng/mL hyper IL6 and 5 × 109 decoy EVs for 5 days. Scale bars, 500 μm. These micrographs are representative of three biological replicates for each group.
(H) Mature osteoclasts with three or more nuclei per well (n = 3) were counted.
The data are representative of two (B–F) or three (G) independent experiments with biologically independent samples. All data are presented as mean ± SEM.
(C and D) Statistical significance was calculated using one-way ANOVA with Tukey’s post hoc test for multiple comparisons.
(F) Statistical significance was calculated using one-way ANOVA with Dunnett’s multiple comparisons test.
(H) Statistical significance was calculated using two-way ANOVA with Tukey’s post hoc test for multiple comparisons.