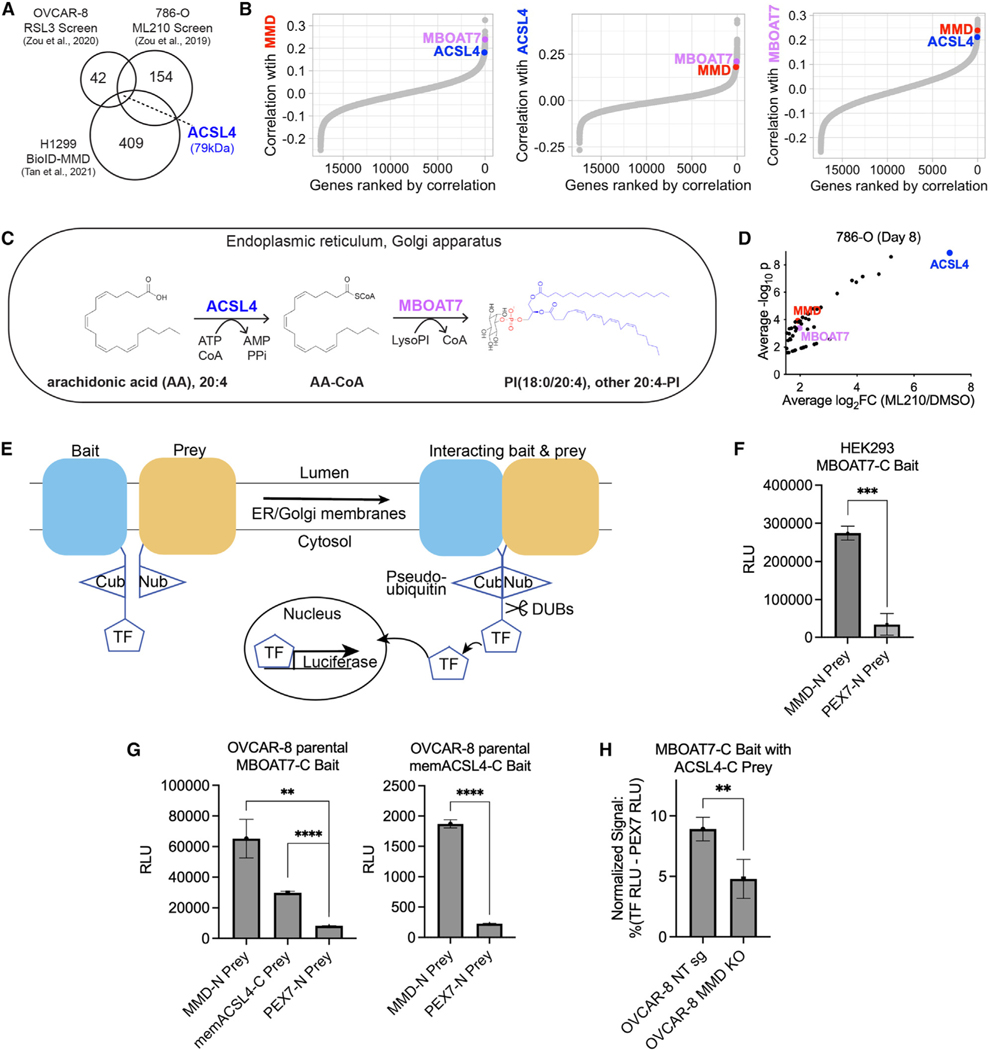
Figure 3. MMD interacts with consecutively acting, membrane-embedded lipid metabolic enzymes ACSL4 and MBOAT7.
See also Figure S3.
(A) Venn diagram showing the intersection of the two published CRISPR screens with a published proximity labeling (BioID) dataset identifying potential interactors of MMD in H1299 cells. Numbers in circles represent total hits in each dataset.
(B) Ordered Pearson correlation coefficients of the effects of loss of MMD (left), ACSL4 (middle), or MBOAT7 (right) with that of loss of every other gene in the genome across all cancer cell lines in DepMap (22Q2). See Table S1 for full ranked lists.
(C) Schematic of the consecutive reactions that ACSL4 and MBOAT7 catalyze to generate PI(18:0/20:4) and other AA-PI.
(D) Volcano plot of hits with average log2fold change ≥1.5 from previously published 786-O CRISPR screen, highlighting MMD, ACSL4, and MBOAT7. Data from Zou et al.12
(E) Schematic of MaMTH. Cub, C-terminal half of ubiquitin; Nub, N-terminal half of ubiquitin; TF, transcription factor; DUBs, deubiquitinase enzymes. Bait andprey are tagged such that the Cub and Nub respectively are cytosol facing. When the bait and prey interact, Cub and Nub are reconstituted as pseudoubiquitin, allowing DUBs to recognize pseudoubiquitin and cleave off the TF. The TF is now free to enter the nucleus and promote expression of the luciferase reporter.
(F) Luciferase activity (relative light units [RLU]) indicating MaMTH reporter activity in HEK293 cells expressing C-terminally tagged MBOAT7 (MBOAT7-C) bait and N-terminally tagged MMD (MMD-N) prey or N-terminally tagged PEX7 (PEX7-N) prey. Bars show mean ± SD of n = 3 biological replicates.
(G) Luciferase activity (RLU) indicating MaMTH reporter activity in OVCAR-8 parental cells. Left: cells expressing MBOAT7-C bait and MMD-N prey, C-terminally tagged membrane-embedded ACSL4 (memACSL4-C) prey, or PEX7-N prey. Right: cells expressing memACSL4-C bait and MMD-N prey or PEX7-N prey. Bars show mean ± SD of n = 3 biological replicates.
(H) Relative MaMTH reporter activity in OVCAR-8 NT sg or MMD KO cells transfected with MBOAT7-C bait and memACSL4-C prey. Normalized signal is expressed as a percentage of signal generated by the positive control (signal in cells transfected with the luciferase-driving TF) minus the signal generated by the negative control (N-terminally tagged PEX7 prey) in each cell line. Bars show mean ± SD of n = 5 biological replicates.
All experimental figure panels are representative of three independent experiments. Statistical analyses by Student’s t test; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.