Figure 6. Intrinsic IFNγ signaling is required for the differentiation of pre-MEMs.
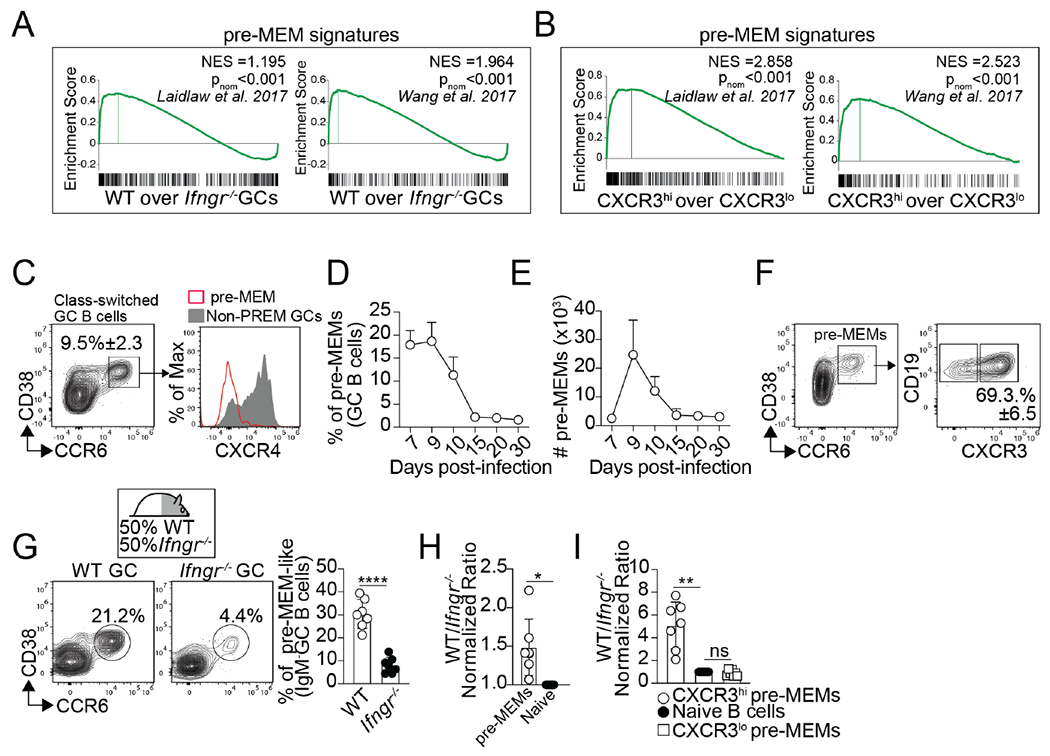
(A) WT and IfngR1−/− GC B cells were sorted from WT/IfngR1−/− BM chimeras on day 12 after PR8 infection and RNA-seq was performed. GSEA for the indicated pre-MEM gene signatures in WT vs. IfngR1−/− GC B cells. Three replicates for each cell type were obtained from three independent experiments. (B) CXCR3hi and CXCR3hi GC B cells were sorted from day 12 PR8-infected B6 mice and RNA-seq was performed. GSEA for the indicated pre-MEM gene signatures in CXCR3hi vs. CXCR3hi GC B cells. Three replicates for each cell type were obtained from three independent experiments. (C-F) B6 were infected with PR8. (C) Gating strategy for the identification of pre-MEMs. Frequency (D) and number (E) of pre-MEMs in the med-LN. (F) Expression of CXCR3 in pre-MEMs on day 10. Plots gated on CD19+CD138−IgD−IgM−FAS+ GC are shown. Data are representative of three independent experiments (n=5 mice/time point). (G) Frequency of pre-MEMs within B6 and IfngR1−/− GC B cells from the med-LN of day 10 PR8-infected WT/IfngR1−/− BM chimeras. (H) Ratio of B6 to IfngR1−/− pre-MEMs. (I) Ratio of B6 to IfngR1−/− CXCR3+ and CXCR3− pre-MEMs. Data are representative of three independent experiments (n=7 mice). P values were determined using a two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
