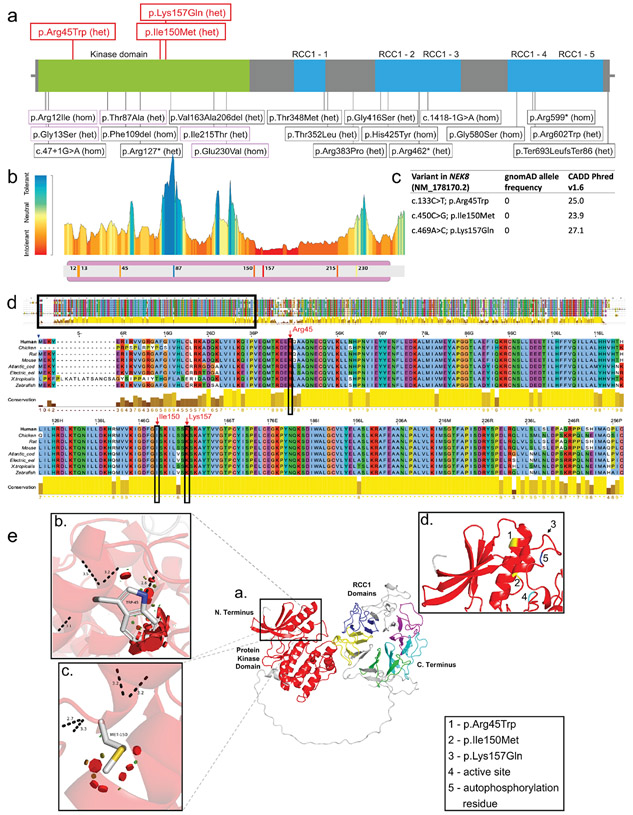
Figure 2. Position of variants and domain organization of the NEK8 protein.
a. In red novel heterozygous variants described in this paper. In black variants previously published in biallelic cases. b. Protein Tolerance Landscape with identified variants and previously reported missense variants in kinase domain highlighted. Color coding indicates predicted impact of variant on protein function with blue =most tolerant, yellow= neutral, red=most detrimental. c. In silico predictions of the novel heterozygous NEK8 variants. d. Multiple sequence alignment of the human NEK8 full protein zoomed in on well conserved kinase domain. The amino acid positions of the heterozygous NEK8 variants are marked by the red arrows. e. Structural modeling of the NEK8 variants. Inset a. The predicted 3-dimentional structural model of human Serine/threonine-protein kinase NEK8 generated using AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk) and UniProtKB (https://www.uniprot.org/uniprot/) with associated codes: AF-Q86SG6-F1 and Q86SG6 respectively. The protein contains a protein kinase domain (red) and five ‘Regulator of Chromosome Condensation 1’ (RCC1) repeat domains. Inset b. Missense SNV c.133C>T p.Arg45Trp modelled with the most probable (26.5% likelihood) rotamer demonstrated. Inset c. Missense SNV c.450C>G p.Ile150Met modelled with the most probable (28.5% likelihood) rotamer demonstrated. Inset d. Positions of both variants in b. and c. highlighted in yellow (and shown as 1. and 2., respectively) to demonstrate their juxtaposition in adjacent alpha-helices. The third variant c.469A>G p.Lys157Gln could not be predicted due to a low confidence score, but appears to be in close proximity to the other variants as highlighted by the arrow (shown as 3.). A known active site (4., proton acceptor) and autophosphorylation residue (shown as 5.) are also in close proximity to the variants.