Figure 6. Dovitinib transcriptionally upregulates IFNG expression by repressing SNAI1/2.
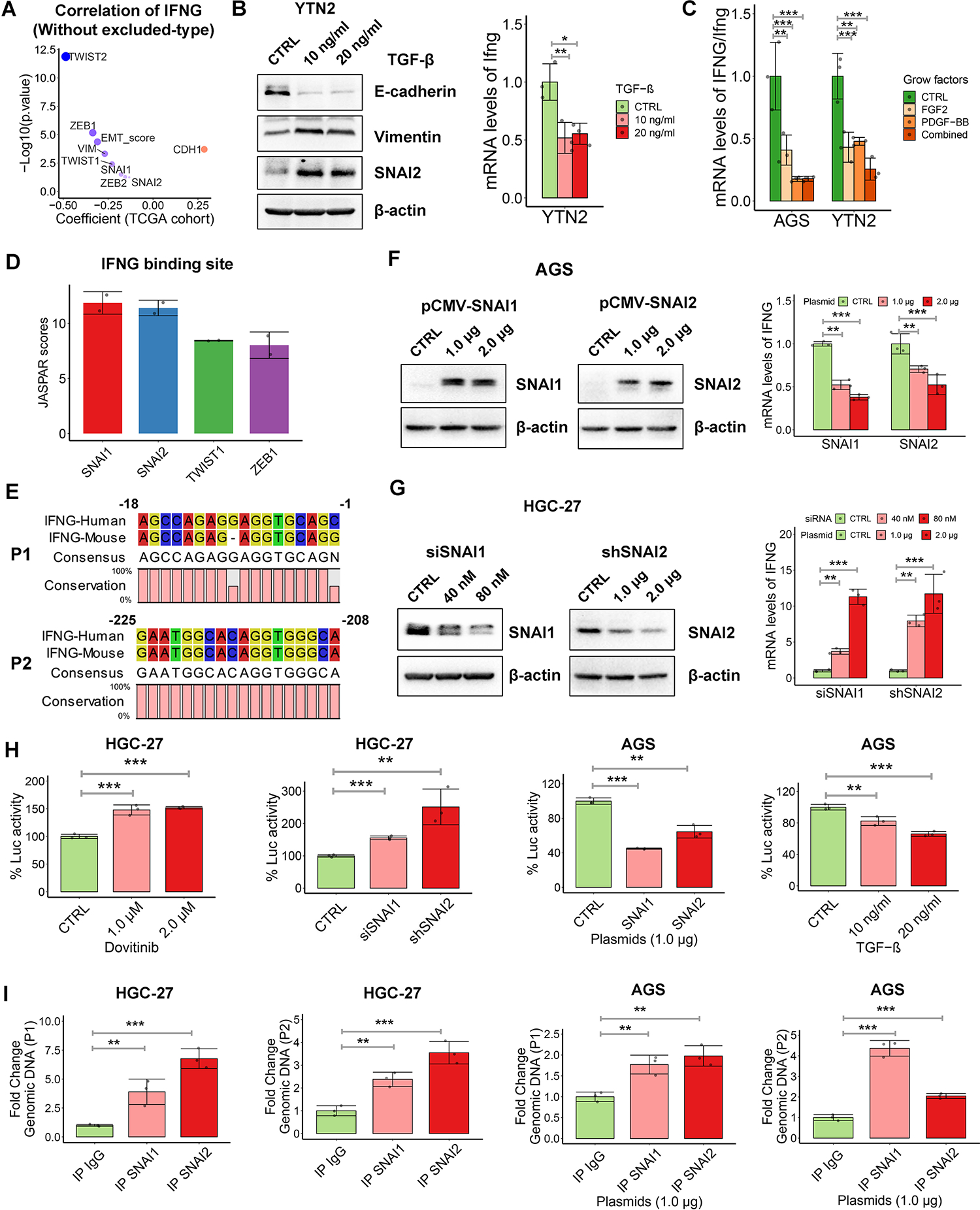
(A) Pearson’s correlation between IFNG expression and EMT score or EMT marker genes expression in the inflamed-type and desert-type of TCGA cohort. (B) Western blots of E-cadherin, Vimentin and SNAI2 in YTN2 cell with the treatment of indicated doses of TGF-β for 3 days (left). qRT-PCR analysis of Ifng in YTN2 cell with treatment of TGF-β (right), (C) qRT-PCR analysis of IFNG/Ifng in gastric cancer cells with treatment of FGF2, PDGF-BB or the combination. (D and E) JASPAR scores (https://jaspar.genereg.net/) of EMT-TFs binding sites (D) and two putative SNAI1/2 binding regions named P1 and P2 (E) in the conserved sequence of IFNG promoters. (F and G) Western blots for SNAI1/2 and qRT-PCR analysis of IFNG in AGS cells with overexpression of SNAI1 or SNAI2 (F), or in HGC-27 cells with knockdown of SNAI1 or SNAI2 (G). (H) IFNG promoter luciferase reporter assays were performed in HGC-27 cells with dovitinib treatment or transient knockdown of SNAI1/2, and in AGS cells with transfection of SNAI1/2 plasmids or TGF-βtreatment. (I) Chromatin immunoprecipitation (ChIP) assays were performed by using SNAI1 or SNAI2 antibodies, followed by qPCR applying primers covering P1 or P2 region. IgG works as non-specific pull-down control. AGS cells were transfected with SNAI1/2 plasmids. In all panels, *p < 0.05, **p < 0.01, ***p < 0.001.
