Figure 2. Fungal pheromone candidates can be identified by a progressive filter for small open-reading frames that are farnesylated and cleaved by a protease associated with mating.
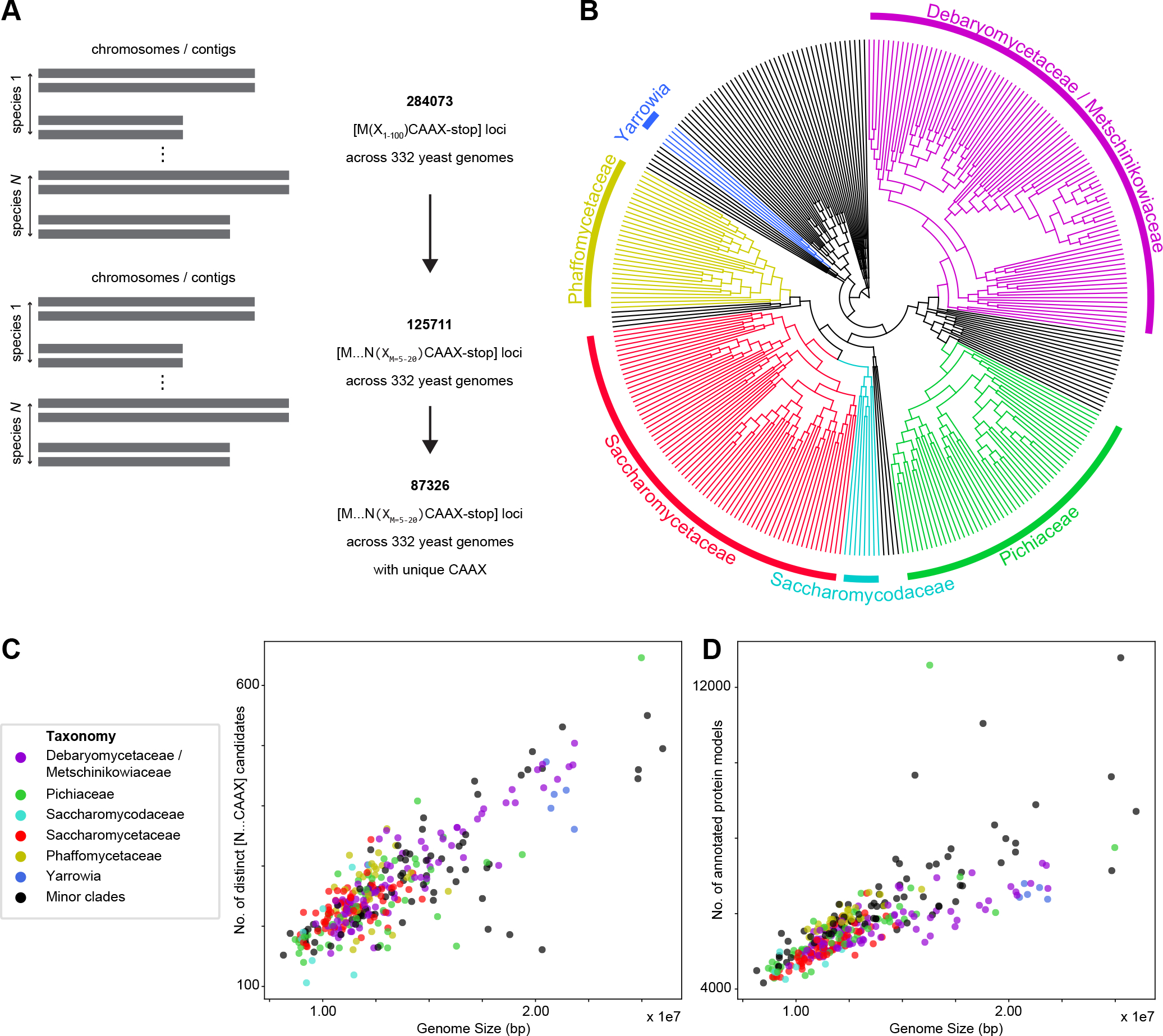
(A) Our algorithm begins by looking for all possible short open-reading frames with C-terminal farnesylation (CAAX-stop) in the 332 available fungal genomes, resulting in 284,073 candidates. We filter again for an in-frame proteolytic-motif asparagine (N) important for the final step of maturation to produce bio-active pheromone. This resulted in 125,711 candidates. Collapsing sets of candidate sequences that result from multiple Start codons upstream of a single CAAX to one sequence from the most upstream Start codon results in 87,326 unique farnesylated loci. (Figure S2E) (B) Phylogenetic tree of all 332 sequenced yeasts across Saccharomycotina, covering clades like Debaryomycetaceae/Metschinikowiaceae, Pichiaceae, Saccharomycodaceae, Saccharomycetaceae, Phaffomycetaceae and Yarrowia that contain species important for both basic biology and industrial production. The tree is based on those yeast genome sequences with estimated relative divergence times27. Selected clades are labeled. (C) Scatter plot showing that the number of unique pheromone candidate loci is linearly correlated with genome size, ranging from 100 to 650 candidates, as expected of a search for all possible pheromone candidates. (D) Scatter plot showing strong correlation between the number of annotated protein-coding genes and genome size, which ranges between 8 and 27 Mbp for the 332 yeast genomes. See also Figure S2.
