Figure 5. All species in Yarrowia clade of yeasts have a homologous farnesylated pheromone that is encoded in multiple copies per genome.
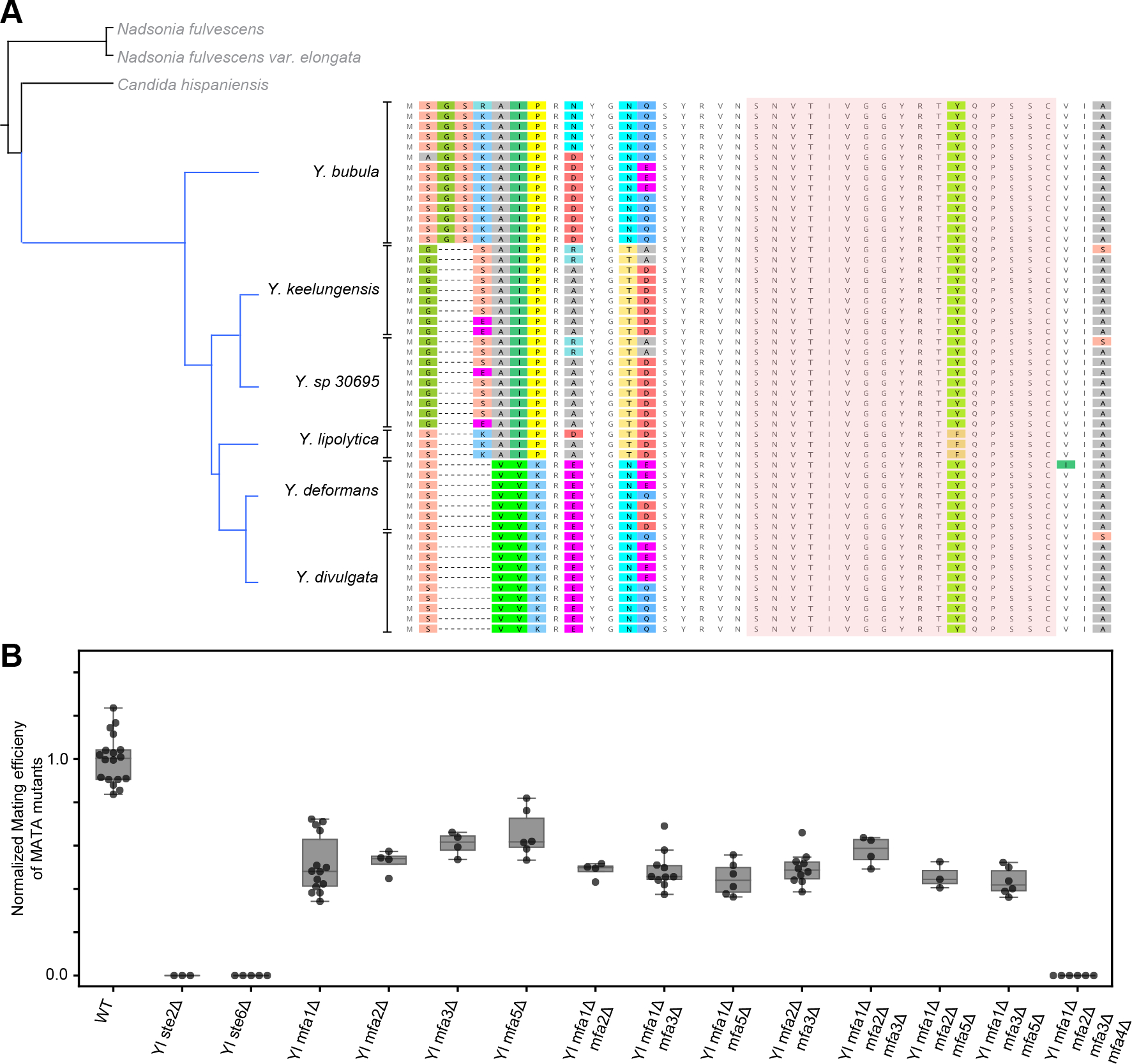
(A) Phylogenetic tree of Yarrowia species (and outgroup in grey) from the set 332 of sequenced yeast of genomes27, along with Y. sp 30695 which is a sister species of Y. keelungensis. Analysis of the genome of Y. sp. 30695 also produced copies of an identical pheromone candidate. The translated ORFs of curated candidates from each species are aligned by the nucleotide sequence of the coding region and ordered according to the phylogenetic relationship between the species. The red shaded region represents the candidate mature pheromone sequence, showing no non-synonymous variation across Yarrowia, except for a conservative change of phenylalanine (F) to tyrosine (Y) in Y. lipolytica. (B) The mating efficiency of MATA haploid derivatives of Y. lipolytica with combinations of the four pheromone loci deleted was evaluated using a semi-quantitative mating protocol. Measurements for each genotype are represented as a group of at least 2 biological replicates each with 2 technical replicate measurements. Single- double- and triple- mutants of YlMFA genes show reduced mating, but only the quadruple deletion of all pheromone loci is deficient in mating to a comparable degree as the receptor (Ylste2Δ) and pheromone exporter (Ylste6Δ) deleted strains. See also Figure S5 and Table S3, S4.
