Figure 1. BPNet predicts a hierarchical relationship between Zelda and patterning TFs in the early Drosophila embryo.
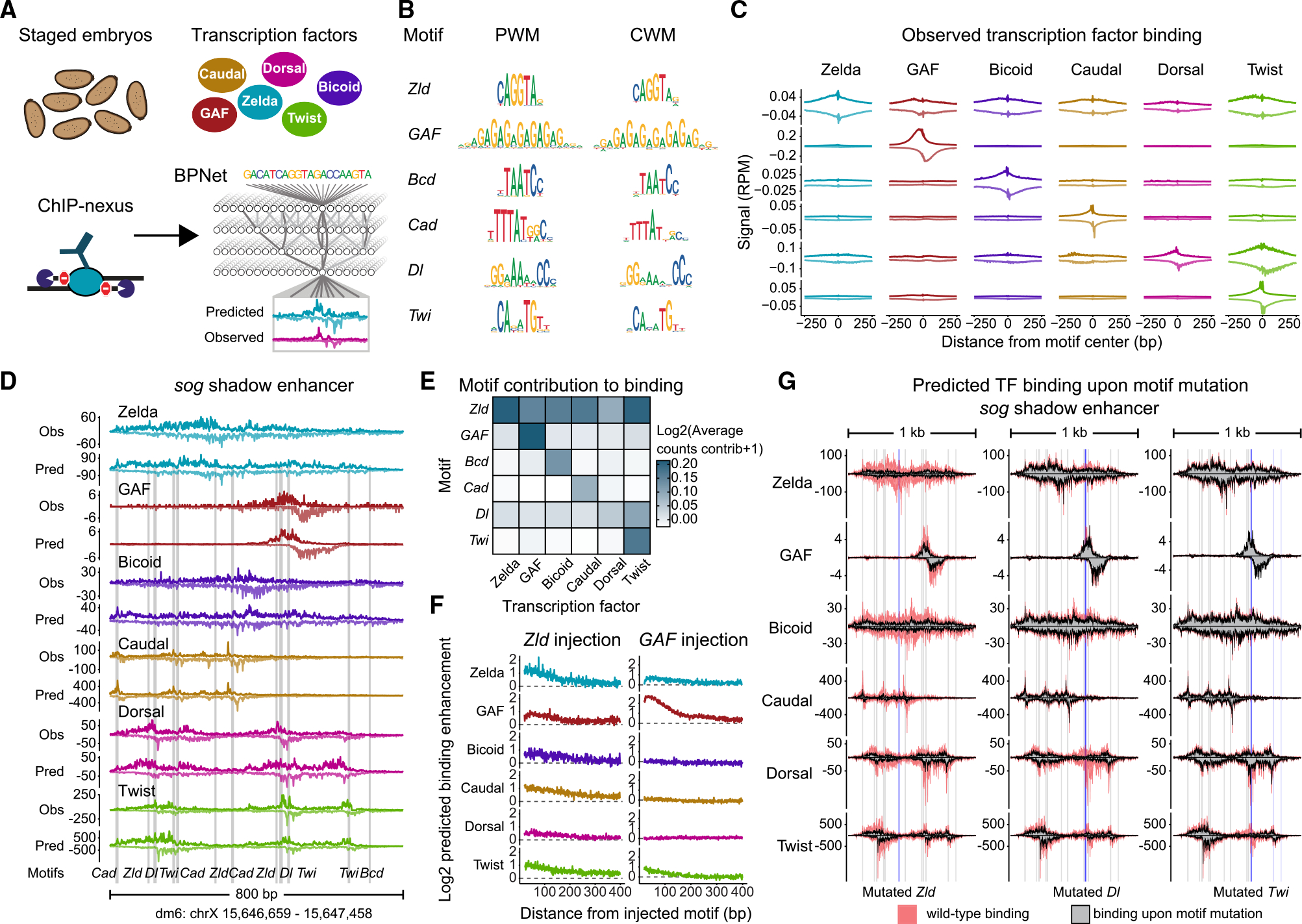
(A) ChIP-nexus produced high-resolution, strand-specific binding of Zelda (Zld), GAGA factor (GAF), Bicoid (Bcd), Caudal (Cad), Dorsal (Dl), and Twist (Twi) in stage 5 embryos. A multi-task BPNet model was trained to predict TF binding from DNA sequence. See also Figures S1A and S2A–S2B.
(B) Identified motifs are shown as a frequency-based position weight matrix (PWM) and as a contribution weight matrix (CWM), which are highly similar for all TFs. See also Figure S2C.
(C) Average ChIP-nexus TF binding footprints show that motifs directly bound by a TF have sharp footprints. Strand-specific data (+ strand on top; − strand at bottom) in reads per million (RPM) were averaged centered on each motif.
(D) BPNet’s predictive accuracy illustrated at the sog shadow enhancer, which was withheld during training. Observed (Obs) ChIP-nexus data are shown above the BPNet-predicted (Pred) data. Motifs contributing to the predictions are found below. Additional enhancers are provided in Figure S3.
(E) The average counts contribution score for all mapped motifs toward the binding of each TF reveals that the Zelda motif contributes to the binding of all TFs, but not vice versa, indicating a hierarchical relationship. Darker colors indicate that a motif (y axis) has a higher contribution score (shown on log scale) to the binding of a TF (x axis).
(F) In silico injections of motifs into randomized sequences confirm that the Zelda motif is predicted to boost the binding of all TFs, while the GAF motif boosts only GAF’s binding. TF binding was predicted by BPNet when each motif was alone and when a Zelda motif (left), or a GAF motif (right), was injected at a given distance, up to 400 bp away (x axis). The average fold-change binding enhancement in the presence of Zelda/GAF is shown on the y axis.
(G) When mutating a Zelda motif in the sog shadow enhancer, BPNet predicts reduced binding of all TFs, while mutating a Dorsal motif has a smaller but notable effect. Predicted binding at the wild-type sequence (red) is overlaid with the predicted binding when individual motifs are computationally mutated (gray). Blue bars highlight the mutated motifs; gray bars are all other mapped motifs. See also Figure S3.
