Figure 5. Patterning transcription factors increase chromatin accessibility through transcriptional activation.
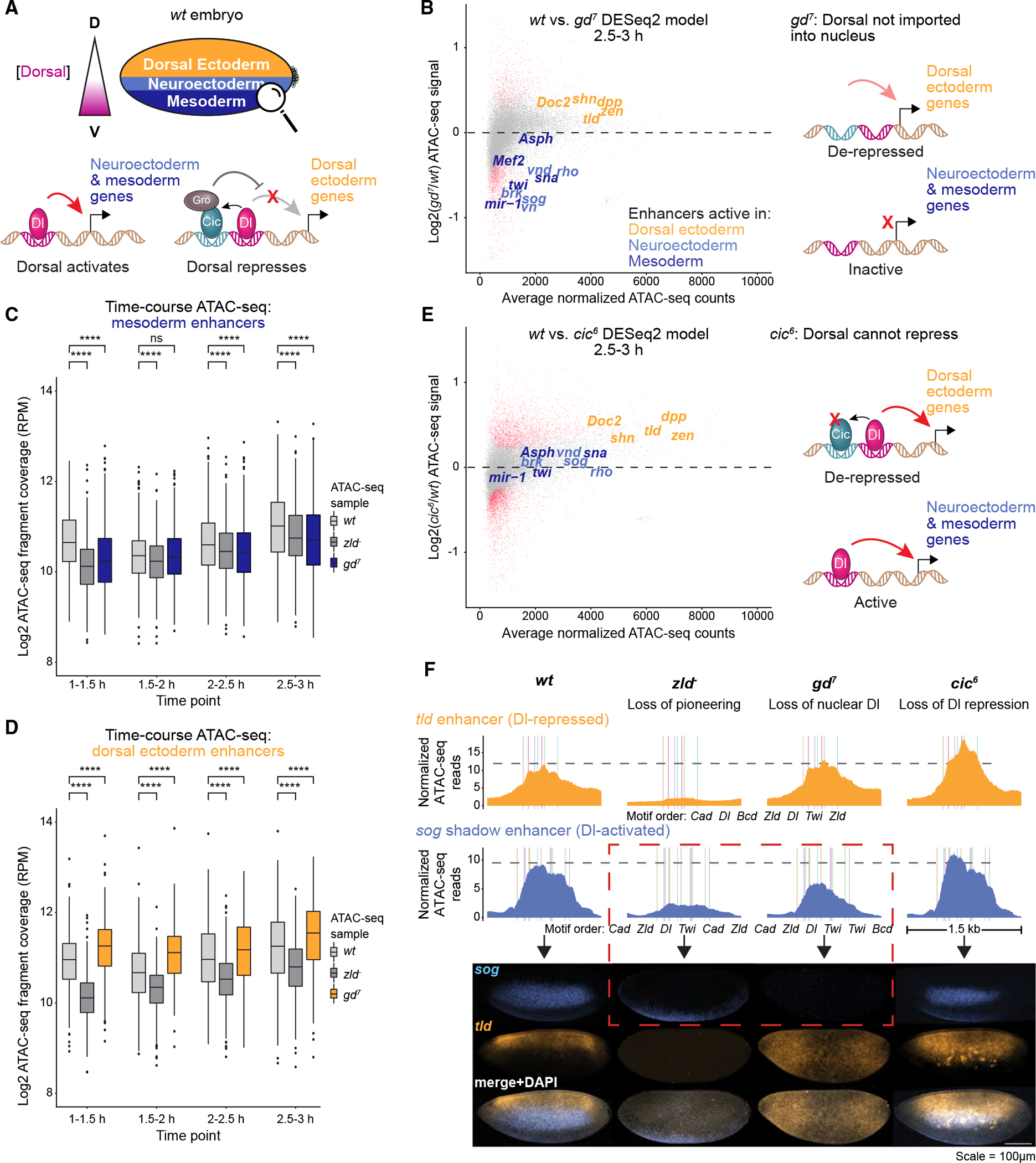
(A) Dorsoventral patterning in the early Drosophila embryo occurs through a nuclear concentration gradient of the Dorsal TF, which activates mesodermal and neuroectodermal target genes but represses dorsal ectodermal genes. Dorsal repression occurs through Capicua, whose binding at these regions depends on Dorsal and which recruits the co-repressor Groucho.
(B) In embryos lacking nuclear Dorsal (gd7), chromatin accessibility is specifically reduced at Dorsal-activated enhancers but not at Dorsal-repressed enhancers. Differential accessibility was calculated between wt and gd7 embryos for all time points and the MA plot for the 2.5–3 h AEL time point is shown. Red dots represent statistically significant differences (false discovery rate [FDR] = 0.05). Known dorsoventral enhancers are colored by the tissue type in which they are active. See also Figures S1F and S6A–S6B.
(C) Mesoderm enhancers, as characterized previously108 (n = 416), have significantly reduced chromatin accessibility in gd7 embryos when they are inactive (Wilcoxon rank-sum tests, four asterisks: p < 0.0001). Normalized ATAC-seq fragment coverage was calculated across 1 kb centered on each enhancer. See also Figure S6C.
(D) Dorsal ectoderm enhancers108 (n = 380) gain chromatin accessibility in gd7 embryos where they are not repressed by Dorsal.
(E) In cic6 embryos, where Capicua’s interaction with Groucho is abrogated and Dorsal can no longer repress, chromatin accessibility is increased at Dorsal-repressed enhancers. Differential accessibility analysis between wt and cic6 embryos was performed as in (B). See also Figures S1G and S6D–S6E.
(F) Chromatin accessibility and target gene activation do not always correlate (dashed red box). ATAC-seq data at a Dorsal-repressed enhancer (tld) and Dorsal-activated enhancer (sog shadow) upon loss of Zelda (zld−), nuclear Dorsal (gd7), and Dorsal-mediated repression (cic6) are shown on top as normalized ATAC-seq fragment coverage from the 2.5–3 h AEL time point across 1.5 kb windows: dm6 coordinates chr3R:24,748,748–24,750,248 (tld) and chrX:15,646,300–15,647,800 (sog shadow). The wt ATAC-seq maximum value is marked as a dotted gray line. Colored bars are BPNet-mapped motifs listed below. Multiplexed hybridization chain reaction experiments show sog and tld expression in stage 5 wt, zld−, gd7, and cic6 mutant embryos (scale is 100 um). Note that sog expression is partially reduced upon loss of Zelda’s pioneering, but completely gone upon loss of Dorsal. Meanwhile, tld expression is ablated in the absence of Zelda but expands upon loss of Dorsal or Dorsal-mediated repression. See also Figures S6F–S6G.
