Figure 1: Midbodies and midbody remnants are sites of RNA storage. MKLP1 and ARC are necessary for mRNA localization and maintenance in the dark zone.
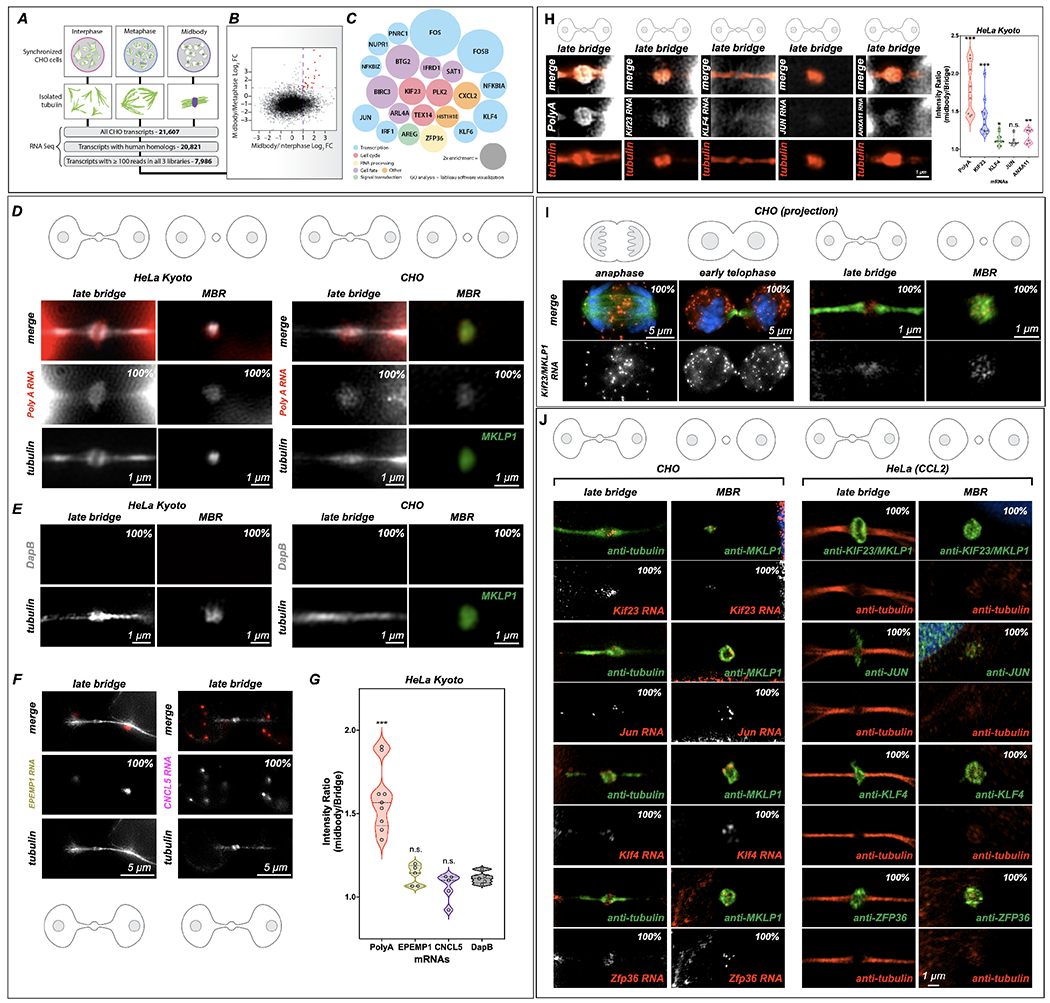
(A-C) RNA-Seq analysis of the MB transcriptome. mRNA was sequenced from three stages of the cell cycle: interphase, metaphase, and late cytokinesis (or “MB stage”). Tubulin structures were purified, and associated RNAs were isolated and analyzed by RNA-Seq. Of 21,607 distinct CHO transcripts identified, 20,821 could be annotated by gene ontology. Of those, 7,986 had ≥100 reads in all cell cycle stages and were further analyzed as plotted in (B). Raw data can be found in Supp.Tables 1–4.
(B) Transcripts with ≥100 reads in all three populations were compared and plotted based on their log2 enrichment scores (RPKM/RPKM). Dotted lines at x = 1 and y = 1 indicate minimum values for 2-fold enrichment. The 22 transcripts enriched in the MB relative to both interphase and metaphase are highlighted in red.
(C) Enrichment score (relative diameter) and gene ontology groups of the 22 MB-enriched transcripts; colors correspond to gene ontology biological process terms (Fig. 1; see also Supp. Tables 1–4).
(D) Single-molecule RNAscope (RNA in situ) hybridization revealed mRNA enrichment in the MB and released MBRs. PolyA-positive mRNAs (red) localized to mitotic MBs and post-mitotic MBRs in both CHO and HeLa Kyoto cells, in contrast with the bacterial DapB negative control (E).
(E) The bacterial DapE was not found at the midbody in HeLa Kyoto or CHO cells midbodies or MBRs.
(F-G) Two mRNAs. EPEMP1 and CNCL5, identified from interphase enriched RNAseq data (see Table 4) were not enriched in the midbody. *Significance was determined and denoted by * or n.s. n.s denotes not significant.
(H) Localization of PolyA, MKLP1, KLF4, JUN and ANXA11 to the midbody in HeLa Kyoto cells. Here we observed an enrichment of PolyA and MKLP1 RNAs but less so the other transcription factors, KLF4 and JUN, and ANXA11 (plot). *Significance was determined by comparing data to DapB 1E. n.s. denotes not significant. See S2C for CHO cell RNAscope quantification for similar probes.
(I) mRNA encoding Kif23, an MB-resident kinesin required for abscission, localized to the spindle overlap from anaphase through abscission; however, in early telophase, Kif23/MKLP1 was also found in the cytoplasm in distinct puncta as well as at the MB dark zone. In late telophase (or G1), puncta were found in the dark zone but were also highly enriched in cell bodies; the released MBR contained Kif23/MKLP1 NA molecules; tubulin is shown in green.
(J) mRNAs identified as MB-enriched by RNA-Seq co-localized to the MB and MBR in CHO cells. In HeLa cells, their complementary proteins were localized to the dark zone and the MBR. RNAscope experiments demonstrated that four mRNAs (Kif23, Jun, Klf4, and Zfp36) localized within the MB matrix, or alpha-tubulin-free zone, of the mitotic MB during G1, and post-mitotically in the MBRs. See S2C for CHO cell RNAscope quantification for similar probes. *Significance was determined by comparing data to DapB 1E. Proteins encoded by these transcripts similarly localized to mitotic MBs and post-mitotic MBRs in HeLa cells.
All data were done in triplicate and quantifications are noted on each figure at a minimum of n=10 for each stage.
