Figure 5: CRISPR-Cas9 Cnr1 deletion from vmPFC→BLA not dmPFC→BLA neurons impairs extinction.
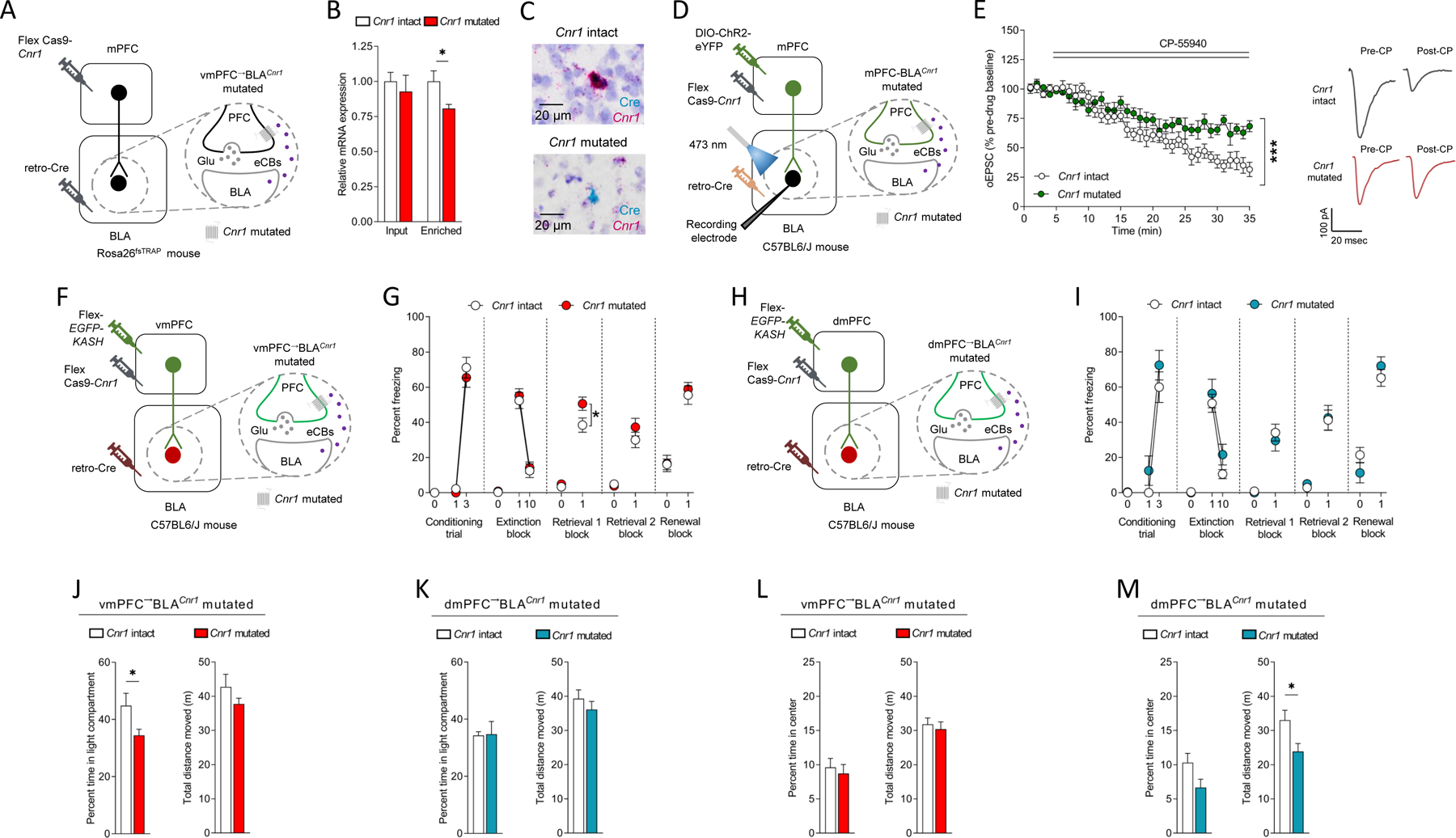
(A-C) Viral strategy for vmPFC→BLA CRISPR-Cas9 Cnr1 mutation in Rosa26fsTRAP mice (A). Less GFP-enriched purified RNA (not GFP-unbound input RNA) expression in mutated versus intact (unpaired t-test: t(11)=2.543, P=0.0273, ηp2=0.37, intact n=6/mutated n=7) (B). Example images from combined BaseScope in situ hybridization and immunohistochemistry showing Cnr1 mRNA loss in Cre-expressing BLA-projecting vmPFC neurons in mutated (not intact) (C).
(D,E) In vitro recordings of CP55–940 (CP, synthetic cannabinoid, CB1R agonist) on ChR2-mediated optically-evoked EPSCs at vmPFC→BLA synapses (D) showing lesser effect in mutated versus intact (ANOVA time x group: F(34,408=4.208, P<0.0001, ηp2=0.07, n=3 mice/6 cells intact/n=3 mice/6 cells mutated), with example traces (E).
(F,G) vmPFC→BLA Cnr1 mutation (F). Freezing increased over conditioning trials (RM-ANOVA: F(1,18)=279.100, P<0.0001, ηp2=0.88) and decreased over extinction trial-blocks (RM-ANOVA: F(1,18)=124.400, P<0.0001, ηp2=0.74). Less freezing on extinction retrieval 1 in mutated versus intact (RM-ANOVA F(1,18)=6.338, P=0.0215, ηp2=0.11) (intact n=9, mutated n=11) (G).
(H,I) dmPFC→BLA Cnr1 mutation (H). Freezing increased over conditioning trials (RM-ANOVA: F(1,13)=87.360, P<0.0001, ηp2=0.69), decreased over extinction trial-blocks (RM-ANOVA: F(1,13)=45.370, P<0.0001, ηp2=0.58) and was similar in mutated and intact across test-phases (intact n=7, mutated n=8) (I).
(J,K) Less light/dark test compartment time in vmPFC→BLA Cnr1 mutated (J) (unpaired t-test: t(18)=2.307, P=0.0331, ηp2=0.23, intact n=9/mutated n=11), not dmPFC→BLA Cnr1 mutated (intact n=8/mutated n=8) (K), versus intact.
(L,M) Less novel open field distance moved (unpaired t-test: t(14)=2.467, P=0.0272, ηp2=0.30, intact n=8/mutated n=8) in dmPFC→BLA Cnr1 mutated (M), not vmPFC→BLA Cnr1 mutated (L), versus intact.
Data are represented as mean±SEM. ***P<0.001, *P<0.05.
