Figure 1. CRISPR-Cas9-mediated AhR gene editing reduces IL-22, IL-17A, and IL-10 production in memory CD4+ T cells.
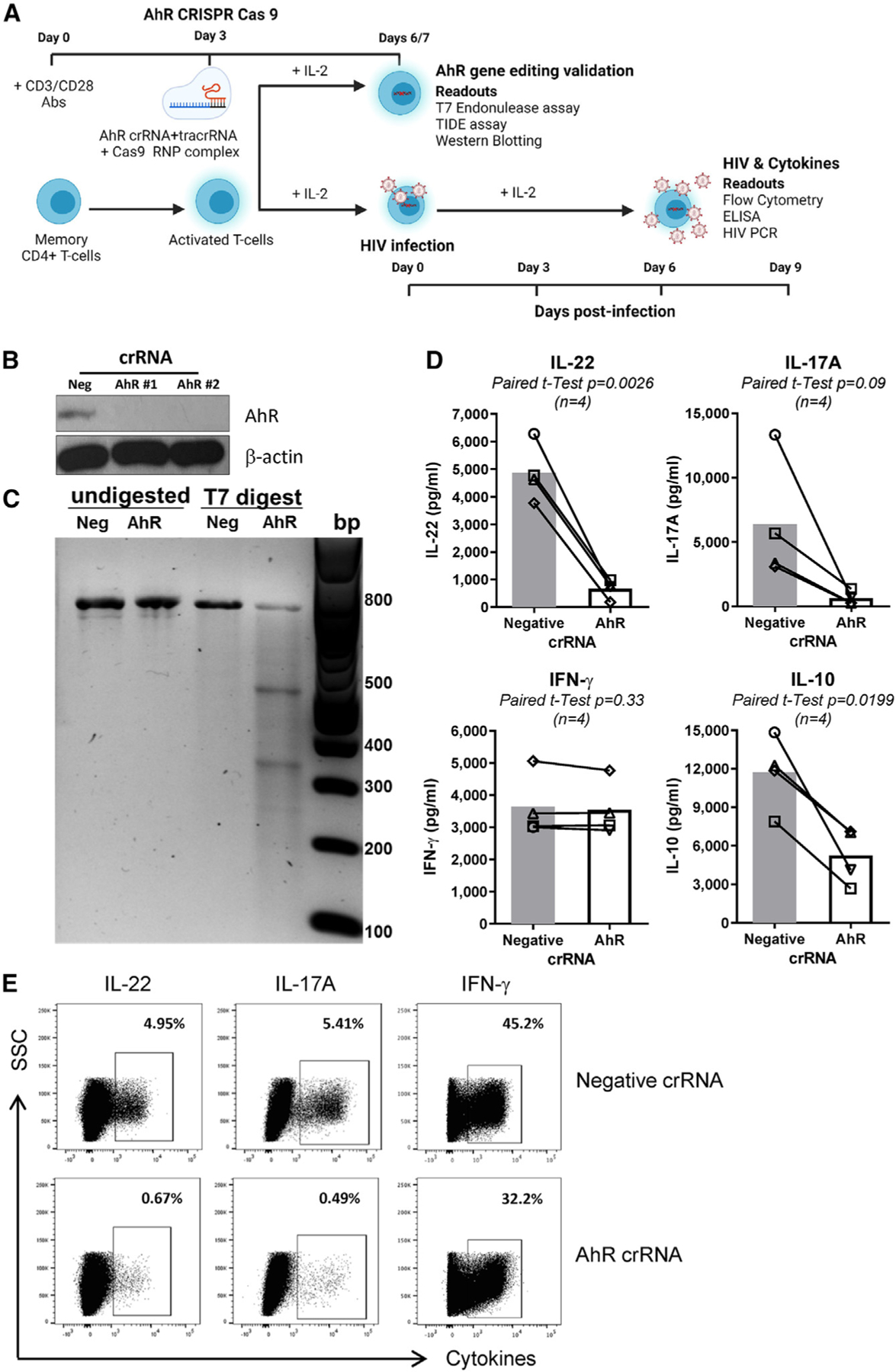
(A) Shown is the experimental flow chart of CRISPR-Cas9 ribonucleoprotein (RNP)-mediated AhR KO. Briefly, memory CD4+ T cells, isolated from peripheral blood mononuclear cells (PBMCs) of HIV-uninfected individuals, were stimulated with CD3/CD28 Abs for 3 days. Cells were electroporated with AhR-targeting (AhR crRNA) or control crRNA (negative crRNA) in the presence of the trans-activating CRISPR RNA (tracrRNA).
(B and C) 3/4 days after electroporation, cells were analyzed by western blotting (B) and the T7 endonuclease assay (C) to determine the efficacy of CRISPR-Cas9-mediated AhR gene editing at protein and DNA levels, respectively.
(D and E) 3 days after electroporation, cells were exposed to HIVTHRO for 3 h, washed, and cultured for 9 days with rhIL-2 (5 ng/mL). At day 9 post-infection, supernatants were collected, and cytokine levels were quantified by ELISA (D). In parallel, cells were stimulated with PMA (50 ng/mL) and ionomycin (1 μg/mL) for 2 h, followed by the addition of brefeldin A (2 μg/mL), and cells were incubated for another 4 h; the intracellular expression of cytokines was measured by flow cytometry (E).
Experiments in (B) and (C) and (D) and (E) were performed with cells from n = 2 and 4 HIV-uninfected individuals, respectively (donors 1–4). Paired t test p values are indicated in (D). Shown are bars that indicate mean values (D).
