Figure 2. Btbd11 contains a PDZ binding motif that interacts with PDZ1,2 of Psd-95.
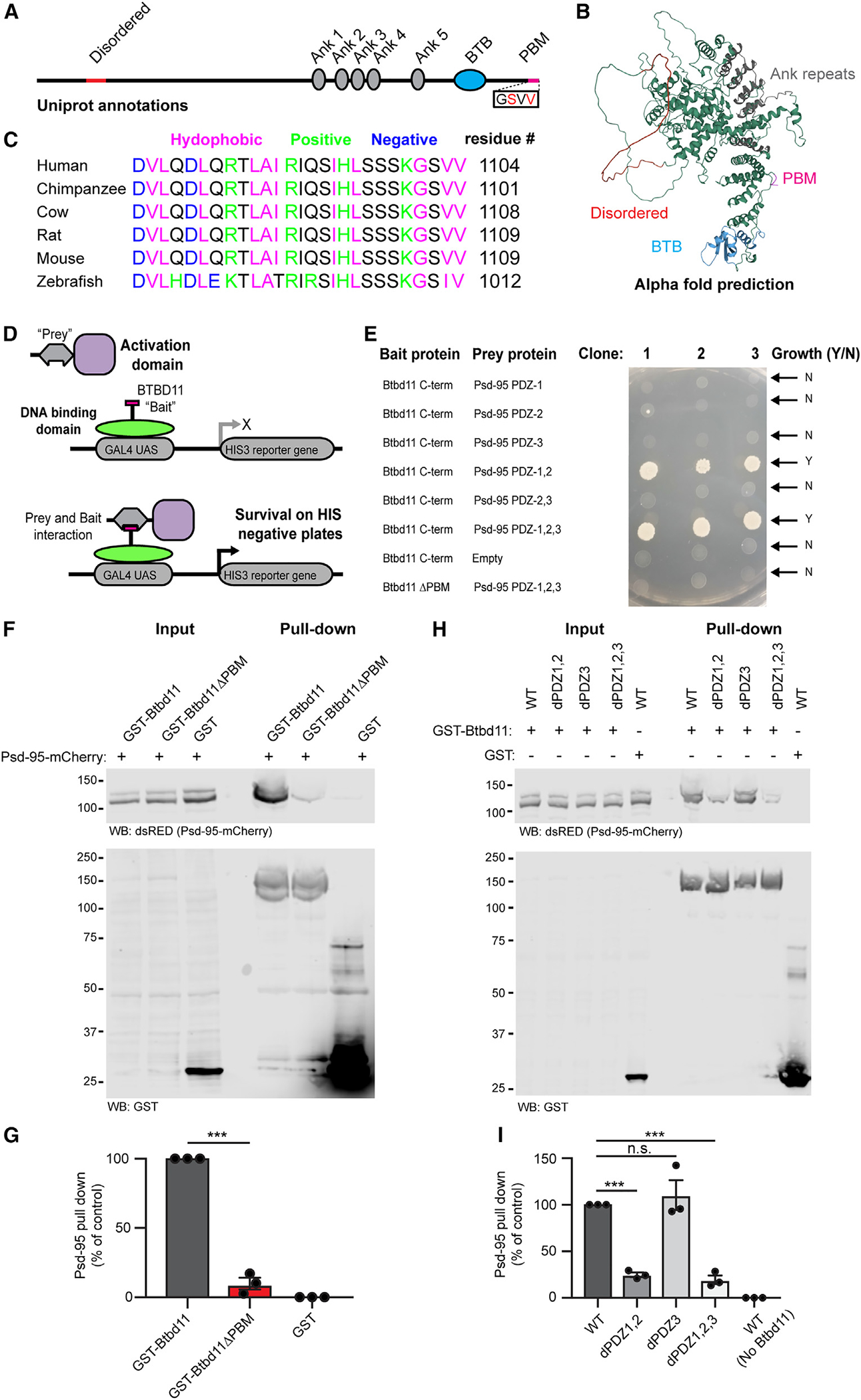
(A) Schematic depiction of Btbd11 with annotations from Uniprot. Red: disordered, gray: ankyrin repeats (Ank), blue: BTB domain (BTB), magenta: PDZ binding motif (PDM).
(B) Predicted structure of Btbd11 from AlphaFold with domains shaded as in (A).
(C) C-terminal region of Btbd11 in different species showing the conservation of the PBM.
(D) Schematic of targeted yeast 2-hybrid experiment to assess binding of Btbd11 with different PDZ domains of Psd-95.
(E) Results from targeted yeast 2-hybrid experiment with growth indicating an interaction between Btbd11 and Psd-95.
(F) GST pull-down experiments evaluating the ability of GST-Btbd11 or a mutant lacking the PBM (GST-Btbd11ΔPBM) to interact with Psd-95-mCherry in HEK cells. GST only was included as a negative control.
(G) Qualification of Psd-95-mCherry pulled down by GST-Btbd11 or GST-Btbd11ΔPBM.
(H) GST pull-down experiments in HEK cells evaluating the ability of GST-Btbd11 to interact with Psd-95-mCherry point mutants designed to disrupt PDZ domain binding.
(I) Quantification of Psd-95-mCherry mutants pulled down by GST-Btbd11. Bars display the mean, and error bars display SEM. ***p < 0.001.
