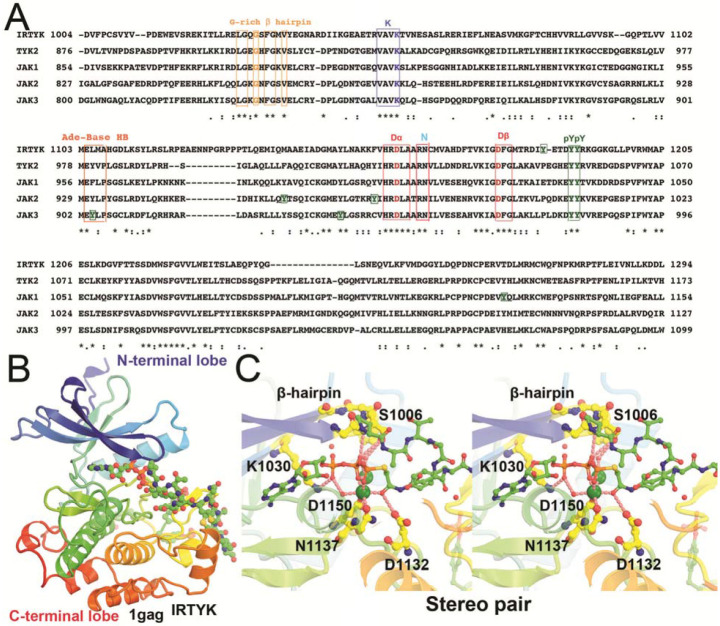
Figure 1.
Kinase domain sequence alignment of IRTYK, TYK2, JAK1 (double-headed Janus kinase), JAK2, and JAK3 in context of the IRTYK complex with bi-substrate analog. (A) Sequence alignment. ATP-interacting motifs are boxed, including the β-hairpin of the GXGXΦG motif (a flip that controls the access of substrate to the γ-phosphate) where Φ is either F or Y, a catalytic triad K, D, and D in the VAVK, HRD, and DFG motifs, respectively, and the adenine base recognition loop of the MEΦXP motif of JAKs. The conserved substrate-docking doublet pY/pY site and other non-conserved single pY sites are also boxed in green. (B) Bilobal kinase domain in complex with the bi-substrate analog in rainbow colors from β-barrel like N-terminal lobe to helical C-terminal lobe. The bi-substrate analog in balls-and-sticks model. (C) Stereodiagram of the close-up view of the kinase active site in which two large green spheres represent two Mg2+ ions.