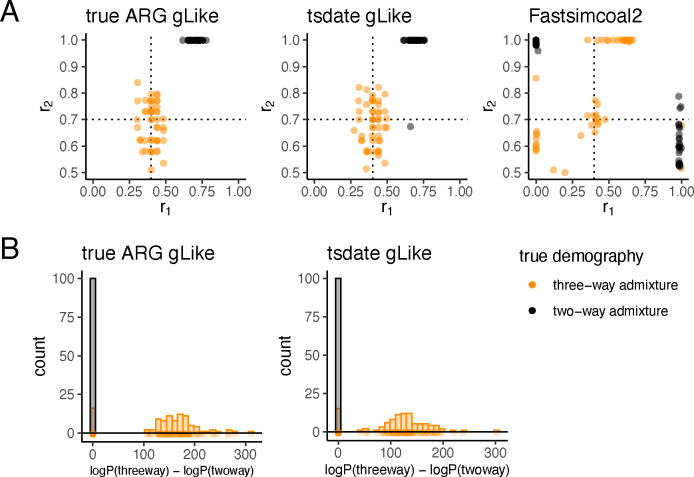
Fig 3. gLike distinguishes three-way admixture from two-way admixture.
True (left) and tsifner+tsdate reconstructed (middle) trees were obtained from simulated three-way (orange, same model as Figure 2) and two-way (grey, was set to 1, removing contribution from population D) admixed populations. (A) gLike was applied assuming a three-way admixture model. The estimated and values in each of 50 independent simulations are shown, dashed lines denote true values of and in three-way admixture simulations. (B) gLike was first applied under a two-way admixture model, then the model is expanded into a three-way admixture and gLike likelihood is optimized while fixing shared parameters between two models (see Methods for technical details). The distributions of log likelihood improvement after model expansion are shown as histogram. Model selection through the Akaike information criterion (AIC) resulted in a classification accuracy of 92%.