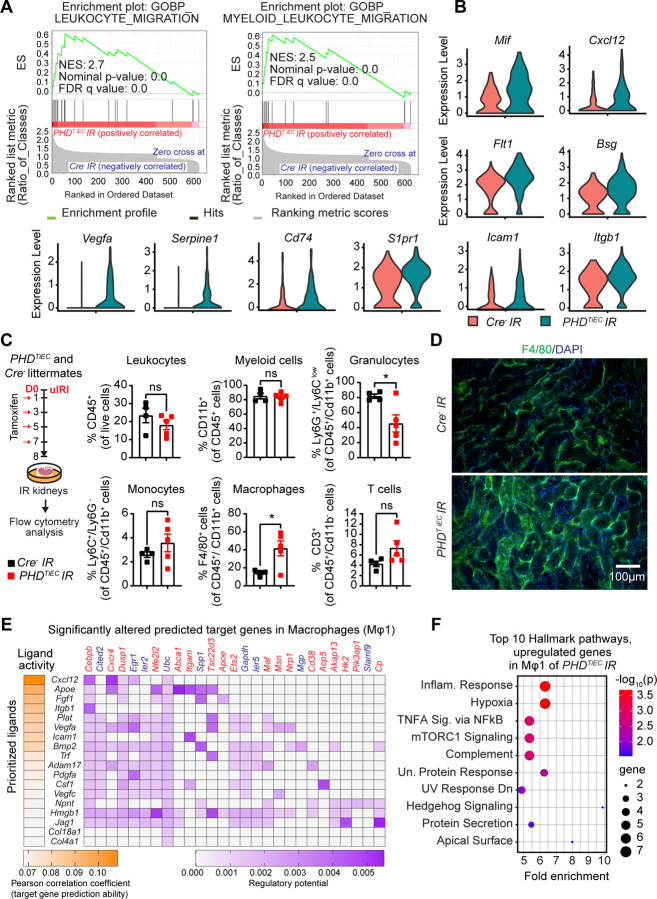
Figure 6. Post-ischemic inactivation of endothelial PHDs induces EC derived pro-inflammatory responses.
(A) GSEA plots show significant enrichment for GO-biological processes (GOBP) of leukocyte migration and myeloid leukocyte migration in mRECs of PHDTiEC post-ischemic kidney compared to control. (B) Violin plots display the expression levels of pro-inflammatory genes associated with core enrichment in GOBP-leukocyte migration and myeloid leukocyte migration in mRECs of PHDTiEC compared to control. (C) Shown is the experimental strategy for flow cytometry analysis. Eight days after uIRI, post-ischemic kidneys from PHDTiEC and Cre− control mice were harvested, and flow cytometry analysis of immune cells was performed (n=4–5). Data are represented as mean ± SEM. Statistics were determined by unpaired t-test with Welch’s correction. (D) Representative images of immunofluorescence staining for F4/80 (green) and nuclear DAPI staining (blue) of day 8 post-ischemic kidneys from PHDTiEC and Cre− control mice. Images were captured using a Nikon Ti2 Widefield fluorescence microscope. Scale bar indicates 100 μm. (E) Shown is NicheNet analysis of mRECs communication with macrophage Mφ1 cluster in day 14 post-IRI kidney of PHDTiEC mutant compared to control. Top prioritized ligands expressed by mRECs (senders) and target genes that are significantly altered (red,upregulated genes; blue,downregulated genes) in the macrophages Mφ1 (receivers). The interaction pairs were derived from the NicheNet data sources and analysis. (F) Hallmark analysis of significantly upregulated genes in Mφ 1 cluster of post-ischemic PHDTiEC kidney compared to Cre− control. Top 10 pathways are shown. *, P <0.05; ns, not statistically significant.