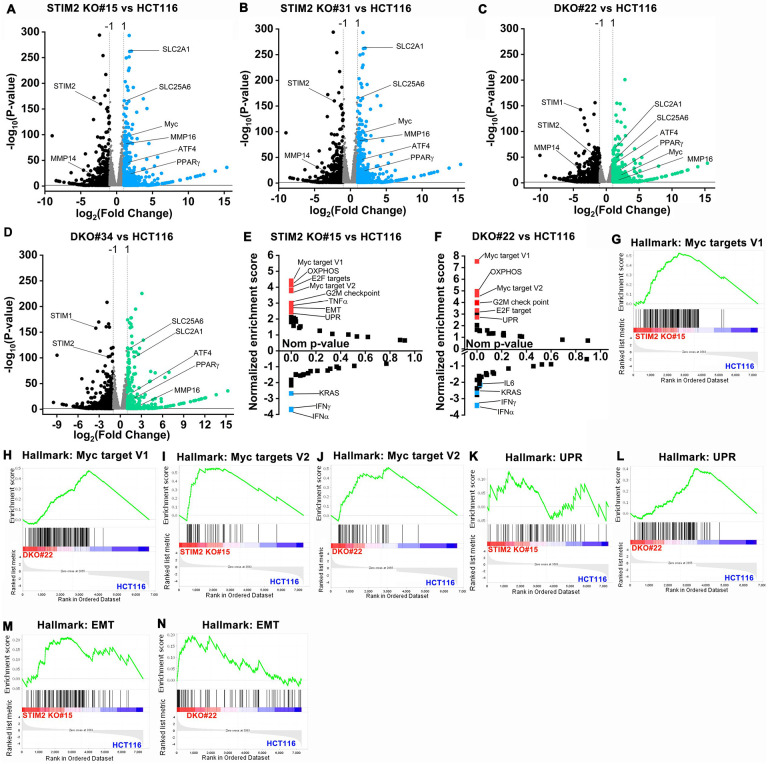
Figure 5. Loss of STIM2 leads to transcriptional reprogramming of CRC cells.
(A-D) Volcano plot showing a comparison of differentially expressed genes between (A) STIM2 KO #15 and HCT116, (B) STIM2 KO #31 and HCT116, (C) DKO #22 and HCT116, and (D) DKO #34 and HCT116. The X-axis represents log2 fold change and Y axis −log10 (P-value). The threshold in the plot corresponds to P- value <0.05 and log 2-fold change <−1.0 or > 1.0. The significantly downregulated genes are represented in black and upregulated in blue or green.
(E, F) The pathway analysis showing plot between normalized enrichment score and nominal p-value. The positive enrichment score represents upregulated pathway, and negative enrichment shows downregulated pathways in (E) STIM2KO #15 vs. HCT116 and (F) DKO #22 vs. HCT116.
(G, H) GSEA analysis between (G) HCT116 and STIM2 KO #15 and (H) HCT116 and DKO #22 show a positive correlation in the enrichment of hallmark of Myc target version 1 genes.
(I, J) GSEA analysis between (I) HCT116 and STIM2 KO #15 and (J) HCT116 and DKO #22 show a positive correlation in the enrichment of hallmark of Myc target version 2 genes.
(K, L) GSEA analysis between (K) HCT116 and STIM2 KO #15 and (L) HCT116 and DKO #22 show a positive correlation in the enrichment of hallmark of unfolded protein response (UPR) genes.
(M, N) GSEA analysis between (M) HCT116 and STIM2 KO #15 and (N) HCT116 and DKO #22 shows a positive correlation in the enrichment of hallmark of epithelial to mesenchymal transition (EMT) genes.