Figure 1.
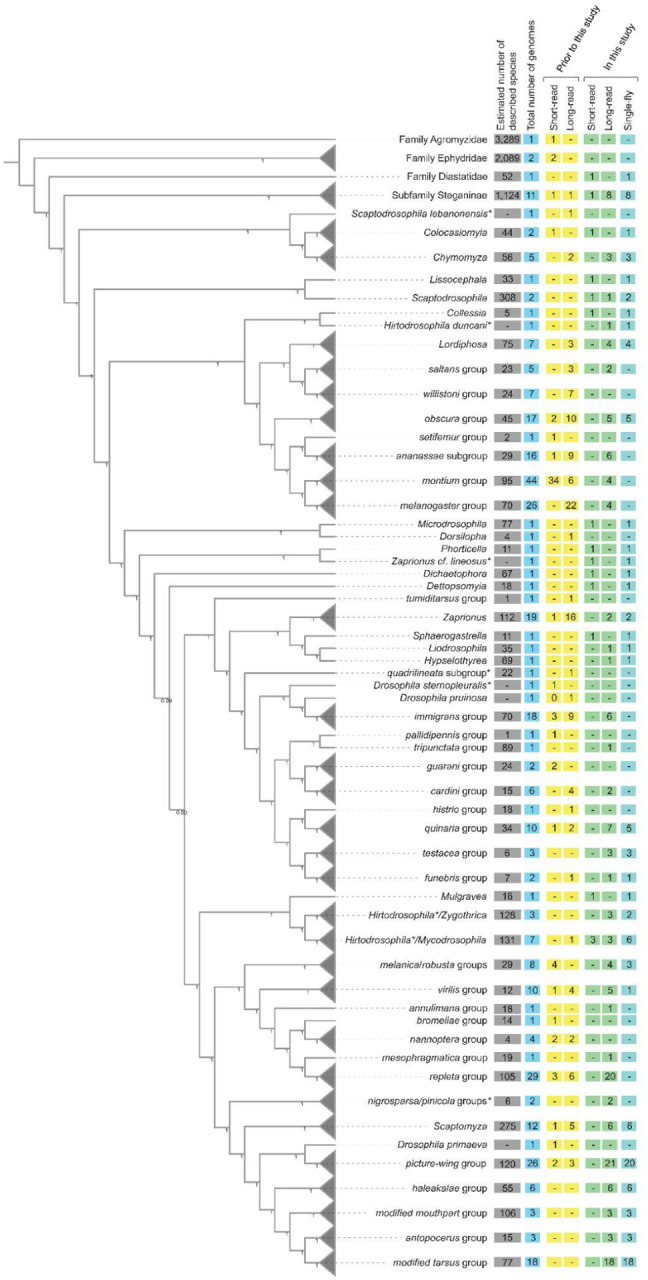
Cladogram of drosophilid species with whole-genome data, with some groups collapsed (gray triangles). Species relationships were inferred from 1,000 orthologs (see Methods). Node values are the local posterior probabilities reported by ASTRAL-MP (Yin et al., 2019). Values in the boxes indicate, as of August 2023, the number of species with whole-genome sequences for each taxon. The count of short-read and long-read datasets, and data available before this study (including Kim et al., 2021) and new genomes presented here are shown separately. *Note that Scaptodrosophila, Hirtodrosophila, Zaprionus, immigrans, and histrio groups are potentially rendered polyphyletic by these samples. The positions of nigrosparsa and pinicola groups are currently considered to be uncertain.
