Figure 2.
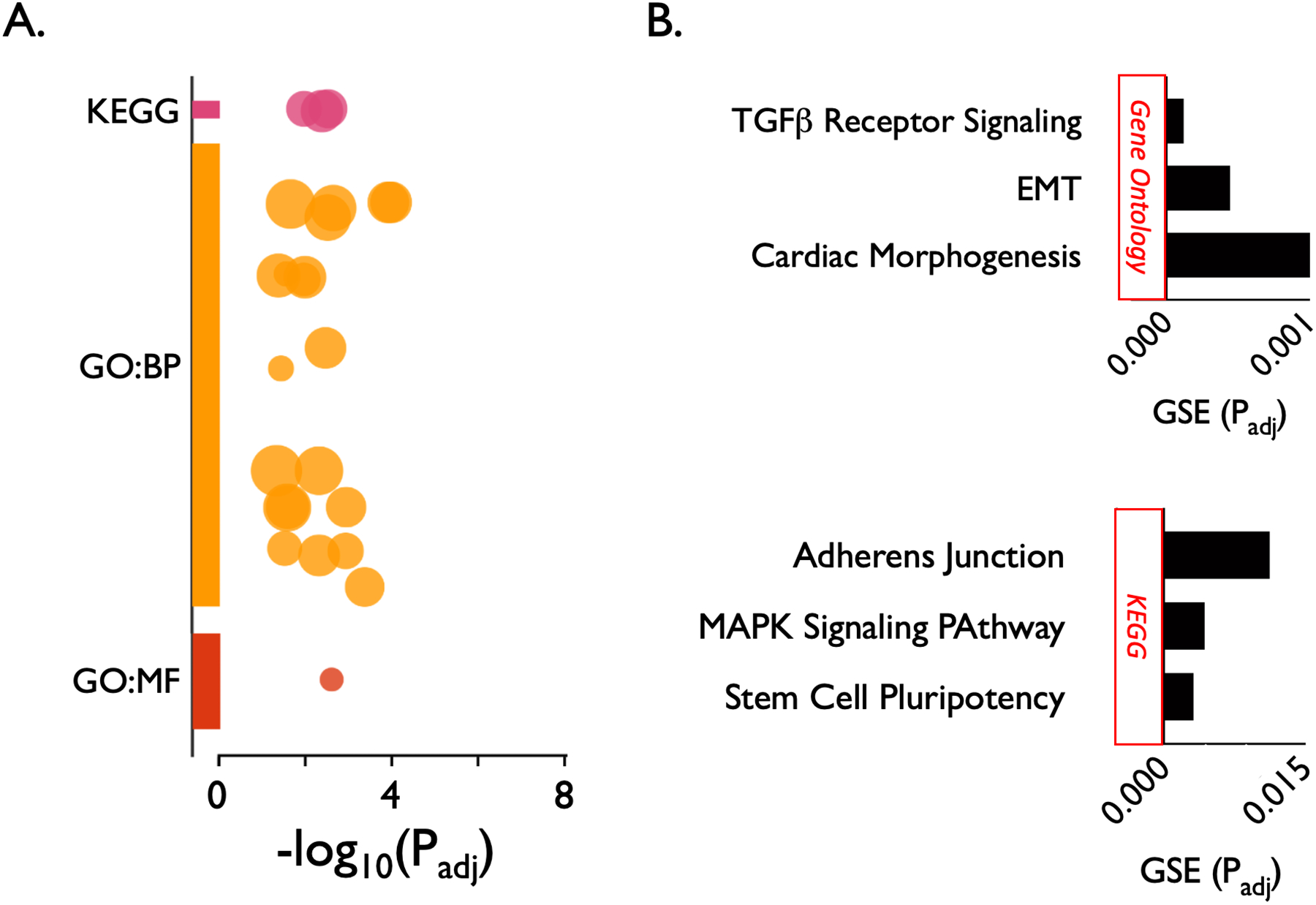
Pathway enrichment analysis of 20 candidate risk genes for subglottic stenosis using g:Profiler. The 24 statistically enriched terms are represented as a Manhattan plot showing each functional term as a circle (A). The top 3 most significantly enriched gene ontology biological process (GO:BP) terms (after collapsing duplicates) were “cellular response to TGF-ß,” “epithelial-to-mesenchymal transition” (EMT), and cardiac “outflow tract morphogenesis.” The single gene ontology molecular function (GO:MF) term was “fibroblast growth factor receptor activity.” The top 3 Kyoto Encyclopedia of Genes and Genomes (KEGG) terms were “signaling pathways regulating pluripotency of stem cells,” “MAPK signaling pathway,” and “adherens junction” (B). X axis represents the adjusted P value of the gene set enrichment (GSE). A complete listing of the 24 significantly enriched biological pathways is listed in Supplemental Table 1.
