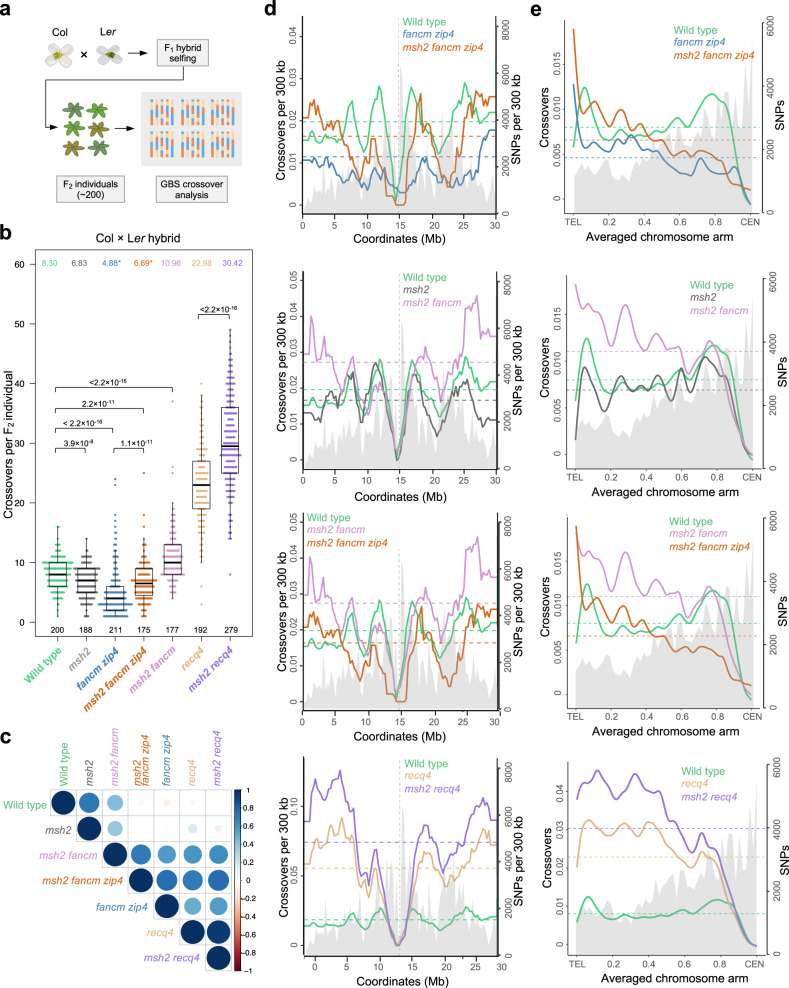
Fig. 2. The effect on MSH2 inactivation on crossover frequency and distribution in different mutant backgrounds.
a Diagram illustrating crossover mapping in F1 plants based on F2 individuals. b The number of crossovers per F2 individual in the indicated populations. For wild type, 200 randomly selected individuals were plotted. Significance was assessed by Kruskal–Wallis H test followed by Mann–Whitney U test with Bonferroni correction. The centre line of a boxplot indicates the mean; the upper and lower bounds indicate the 75th and 25th percentiles, respectively; the whiskers indicate the minimum and maximum. The mean is indicated also on the top. Asterisks by means denote genotypes where crossover numbers are likely to be inflated due to reduced fertility (only gametes with sufficient crossover numbers can form the sequenced F2 generation). The numbers of individuals are indicated below the boxplots. c The correlation coefficient matrices among genome-wide crossover distributions as calculated in 0.3 Mb adjacent windows. d Crossovers per 300 kb per F2 plotted along the Arabidopsis chromosome 1. Mean values are shown by horizontal dashed lines. SNPs per 300 kb are plotted and shaded in grey. The position of centromere is indicated as vertical dashed line. e Data as for (d), but analysing crossovers along proportionally scaled chromosome arms, orientated from telomere (TEL) to centromere (CEN). b–e Data for wild type, msh2 and recq4 from refs. 41,49,66, respectively. Source data are provided as a Source Data file.