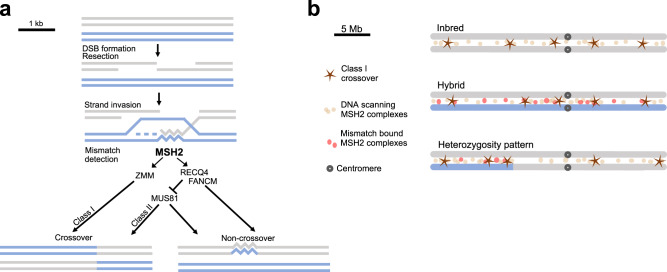
Fig. 6. Models showing the impact of DNA polymorphism on crossover formation during Arabidopsis meiosis.
a Two recombining homologous DNA molecules are depicted in grey and blue, over a region of several kilobases. Following DSB formation on the grey molecule, resection occurs to form 3’-single-stranded DNA. This ssDNA undergoes strand invasion into the homologous molecule, forming a displacement loop. MSH2 heterodimers detect mismatches at the invasion site. Two scenarios are proposed for the situation when mismatches are detected: (i) MSH2 promotes ZMM pathway leading to Class I crossover, or (ii) MSH2 recruits or stimulates DNA helicases, including FANCM and RECQ4, resulting in D-loop displacement and non-crossover repair. In the absence of mismatches or MSH2, MUS81 endonuclease repairs the DSB via Class II crossover or non-crossover. Alternatively, MSH2 can directly stimulate MUS81-dependent crossover formation (not shown). b Two recombining homologues are depicted with grey and blue colours representing sequence divergence. MSH2 complexes scan DNA to detect mismatches in heteroduplexes. In inbreds, when there are no mismatches, the position of crossovers is determined mainly by the chromatin structure. In hybrids, the mismatches along the entire chromosome length trigger a fairly even distribution of mismatch-bound MSH2 complexes, which, combined with interference, also results in Class I crossover placement determined mainly by chromatin. However, the presence of a single heterozygous region on an otherwise homozygous chromosome results in a local concentration of mismatch-bound MSH2 complexes that stimulate Class I crossover in the heterozygous region.