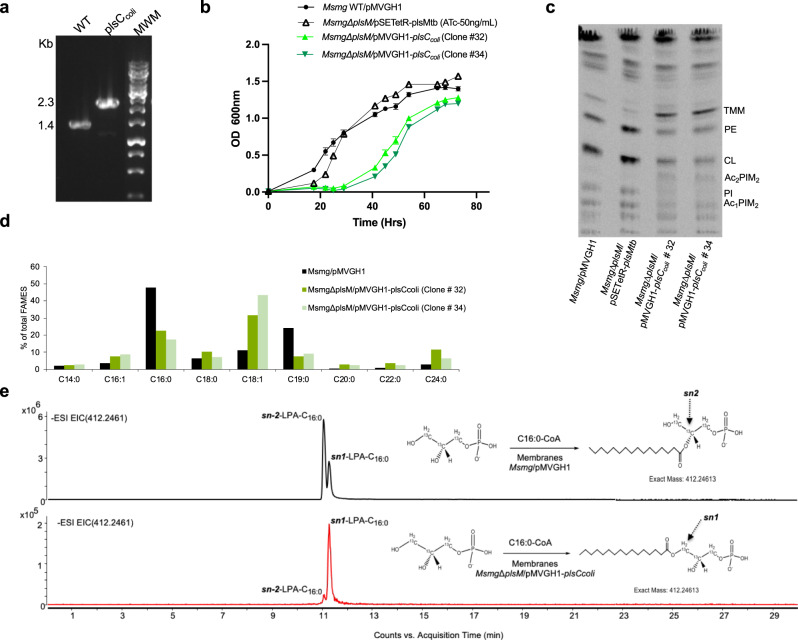
Fig. 2. Effect of replacing PlsM by PlsC from E. coli in Msmg.
a Allelic replacement at the plsM locus of Msmg mutants rescued with plsCcoli was analyzed as in Fig. 1b. b Growth characteristics of WT Msmg mc2155 (black circles), MsmgΔplsM/pSETetR-plsMtb (black triangles), and MsmgΔplsM/pMVGH1-plsCcoli (clones # 32 and 34; light and dark green triangles, respectively) in 7H9-ADC-0.05% tyloxapol at 37 °C (in the presence of 50 ng/mL ATc for MsmgΔplsM/pSETetR-plsMtb). Shown are the means ± SD of three independent growth curves (n = 3 independent biological triplicate) for each strain. c De novo phospholipid synthesis in WT Msmg mc2155, MsmgΔplsM/pMVGH1-plsCcoli (clones # 32 and 34) and MsmgΔplsM/pSETetR-plsMtb grown in 7H9-ADC-0.05% tyloxapol at 37 °C (in the presence of 50 ng/mL ATc for MsmgΔplsM/pSETetR-plsMtb). [1,2-14C]acetic acid was added to bacterial cultures when they reached an OD600 nm of 0.8 after which cultures were incubated for another 8 h at 37 °C with shaking. [1,2-14C]acetic acid-derived lipids were analyzed by TLC in the solvent system CHCl3:CH3OH:H2O (70:20:2 by vol.). The same total counts (dpm) were loaded per lane. Ac2PIM2 are tetraacylated forms of phosphatidyl-myo-inositol dimannosides. Other lipid abbreviations are as in Fig. 1. The results shown are representative of two independent experiments. d Fatty acid composition of WT Msmg mc2155 and MsmgΔplsM/pMVGH1-plsCcoli (two different clones, # 32 and # 34; dark and light green bars, respectively) grown in 7H9-ADC-0.05% tyloxapol at 37 °C (same medium as used in panels b and c). C19:0: tuberculostearic acid. e Extracted ion chromatograms (EICs) showing the lysophosphatidic acid (LPA) products resulting from the incubation of membranes prepared from Msmg/pMVGH1 and MsmgΔplsM/pMVGH1-plsCcoli (clone # 32) with [13C]-G3P and C16:0-CoA. sn-2 palmitoyl transferase activity is dominant in Msmg/pMVGH1 whereas it is barely detectable in MsmgΔplsM/pMVGH1-plsCcoli. C16:0 in the latter strain is essentially transferred to position sn-1 of G3P. Source data for panels (b and d) are provided as a Source Data file.