Figure 2. SVM machine learning-aided identification of a BAG6 degron motif.
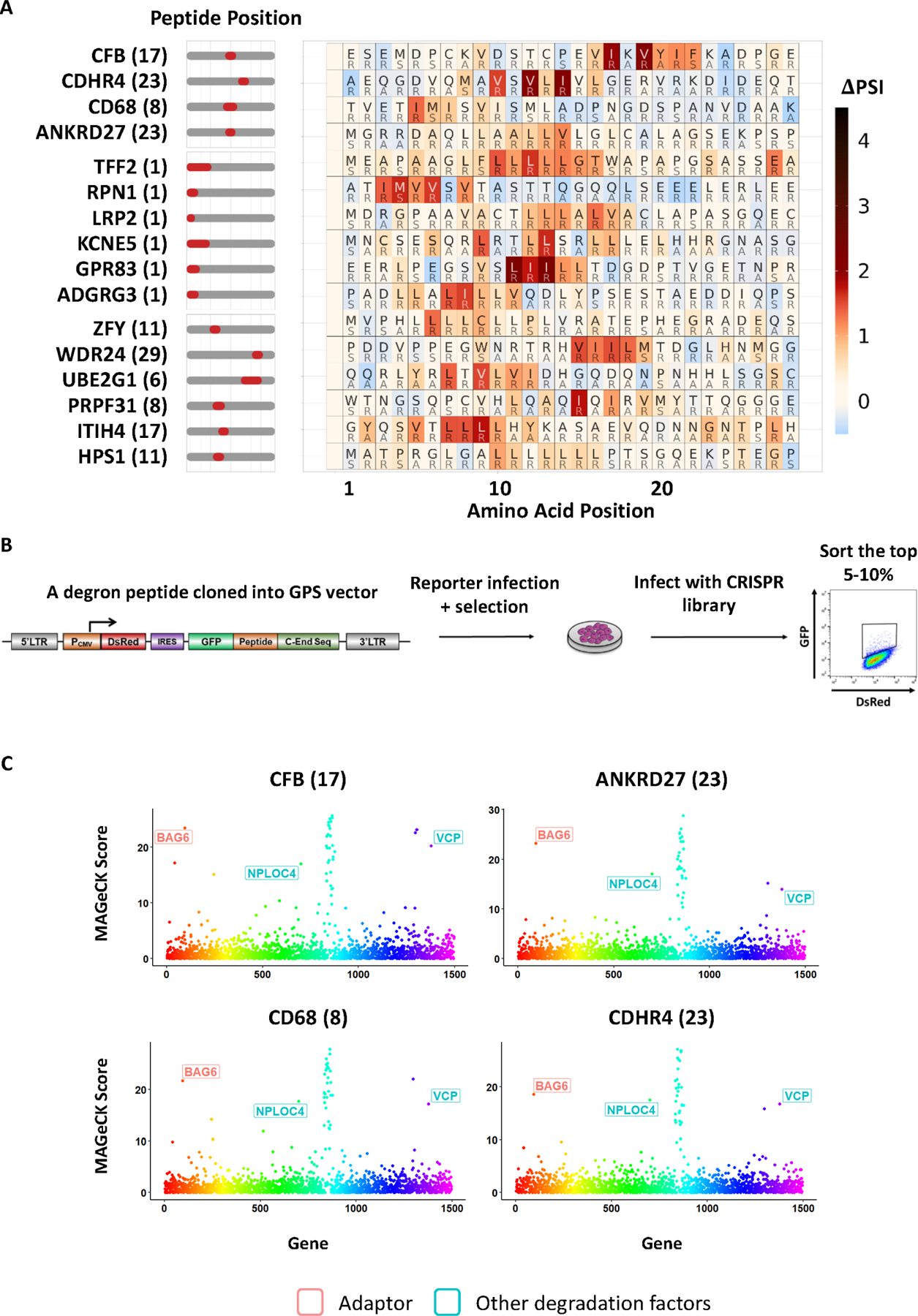
(A) Scanning mutagenesis of non-CRL degron peptides. Peptides in the top group were subjected to CRISPR screens and BAG6 was identified as a destabilizing gene. BAG6-like motifs were found not only in terminal peptides (middle group) deriving from N-terminal signal peptide sequences, but also in internal peptides (bottom group).
(B) Schematic diagram illustrating the workflow of the CRISPR screen designed to identify the genes required for the degradation of each degron peptide.
(C) CRISPR screens of 4 representative degron peptides with motifs described in (A) using a sgRNA lentiviral library of UPS related genes identified BAG6 as a gene required for the degron activity.
