ABSTRACT
Within-host Mycobacterium tuberculosis (Mtb) diversity may detect antibiotic resistance or predict tuberculosis treatment failure and is best captured through sequencing directly from sputum. Here, we compared three sample pre-processing steps for DNA decontamination and studied the yield of a new target enrichment protocol for optimal whole-genome sequencing (WGS) from direct patient samples. Mtb-positive NALC-NaOH-treated patient sputum sediments were pooled, and heat inactivated, split in replicates, and treated by either a wash, DNase I, or benzonase digestion. Levels of contaminating host DNA and target Mtb DNA were assessed by quantitative PCR (qPCR), followed by WGS with and without custom dsDNA target enrichment. The pre-treatment sample has a high host-to-target ratio of DNA (6,168 ± 1,638 host copies/ng to 212.3 ± 59.4 Mtb copies/ng) that significantly decreased with all three treatments. Benzonase treatment resulted in the highest enrichment of Mtb DNA at 100-fold compared with control (3,422 ± 2,162 host copies/ng to 11,721 ± 7,096 Mtb copies/ng). The custom dsDNA probe panel successfully enriched libraries from as little as 0.45 pg of Mtb DNA (100 genome copies). Applied to direct sputum the dsDNA target enrichment panel increased the percent of sequencing reads mapping to the Mtb target for all three pre-processing methods. Comparing the results of the benzonase sample sequenced both with and without enrichment, the percent of sequencing reads mapping to the Mtb increased to 90.95% from 1.18%. We demonstrate a low limit of detection for a new custom dsDNA Mtb target enrichment panel that has a favorable cost profile. The results also demonstrate that pre-processing to remove contaminating extracellular DNA prior to cell lysis and DNA extraction improves the host-to-Mtb DNA ratio but is not adequate to support average coverage WGS without target capture.
KEYWORDS: mycobacteria, target capture, enrichment, direct sputum sequencing
INTRODUCTION
Mycobacterium tuberculosis (Mtb) is the leading infectious pathogen killer globally. Although tuberculosis (TB) incidence has declined over the past decade, the global burden remains at more than 10 million people newly ill with the disease annually (1). DNA sequencing advances over the past decade have enabled the study of Mtb’s genetic epidemiology, while also providing valuable information on within patient Mtb diversity, single-nucleotide polymorphisms (SNPs), and other mutations which can be used to predict drug susceptibility (2, 3). Whole-genome sequencing (WGS) of Mtb is still hindered by the long and cumbersome Mtb culturing process for DNA extraction. Culture can take weeks to months and has the additional limitation of potentially changing the population structure of the original sample due to selection of subpopulations more suited for growth in culture or stochastic purging from population bottlenecks (3, 4).
Sequencing the complete genome directly from clinical specimens would eliminate these issues. Several studies have demonstrated that sequencing from direct patient specimens is possible with varying levels of success (2 – 7). The most successful appraoch to direct sputum sequencing (DSS) has involved the use of target-specific RNA bait probes during library preparation. However, these probes still introduce several limitations, including low uniformity of coverage, the high proportion of duplicate reads generated as a result of PCR steps in both library preparation and enrichment, and technology costs ($110–168 per library and hybridization reaction) (8, 9). Probe-based enrichment can also be limited by target capture specificity, making the approach sensitive to contamination and to the ratio of contaminant to target DNA (2 – 4, 6). A pre-processing step to decrease contaminant DNA burden can thus further boost enrichment of Mtb DNA for WGS (3).
Prior studies have selectively lysed contaminating host and bacterial cells, followed by the depletion of contaminating DNA by enzymes such as DNase, either forgoing target enrichment or only using it in processing samples with low amounts of input mycobacterial DNA (3, 5). Although these studies were successful, this approach is highly dependent on Mtb bacillary load and proportions of contaminants including host cells and DNA as well as host microflora and their DNA (5). There is thus a need for a more consistently effective approach to contaminant depletion and Mtb target capture for DSS.
We postulated that sodium hydroxide (NaOH) decontamination followed by heat killing will lyse most contaminating host cells and non-Mtb bacteria, that Mtb cells are resilient to sodium hydroxide treatment and heat because of Mtb’s thick and lipophilic cell wall. Thus, after cell lysis, the majority of remaining contaminating DNA will be extracellular and thus accessible to enzymatic degradation, while the more resilient Mtb cells remain intact (10, 11). Previous studies have included DNase for depletion of extracellular contamination, but the direct effect on enrichment has not been systematically evaluated. Benzonase is an alternative enzyme for contamination removal that has not previously been evaluated (3, 5). Benzonase breaks down both ssDNA, dsDNA, RNA, and DNA:RNA hybrids, while DNase targets primarily dsDNA with reduced specificity for ssDNA and DNA:RNA hybrids. Benzonase is thus an ideal candidate for extracellular DNA depletion that may provide a more efficient solution than the more frequently used DNase (12 – 14).
In this study, we evaluate benzonase and DNase head to head and compared their effect on enrichment and if enzymatic pre-treatment provides benefit to downstream target capture and enrichment. We also test the Twist (Twist Bioscience, USA) target capture system for Mtb target enrichment. The system is similar or more affordable ($110) than competing platforms and promises more uniform capture through the use of double-stranded DNA (dsDNA) biotin-labeled probes (15). Twist dsDNA probes have been used for Illumina sequencing from a range of starting materials, including respiratory specimens for SARS-CoV-2 WGS, and is hence a good candidate for sputum (15, 16).
MATERIALS AND METHODS
Sample preparation
Limit of detection samples
Sputum sediment samples are expected to contain low amounts of total DNA, of which a small proportion is Mtb DNA. We generated a low limit of detection (LoD) series of spiked samples using a H37Rv (ATCC 27294) reference strain to first assess the Twist kit’s ability to capture, and thus amplify, low amounts of Mtb target and prepare libraries from very low input concentrations (<1 ng). Current manufacturer’s recommendations are to optimally start with 50 ng of input DNA, but feedback from the manufacturers indicate that the kit can still successfully prepare libraries down to 1 ng. Liquid Middlebrook 7H9 medium supplemented with 0.2% glycerol, 0.25% Tween 80 (Sigma, USA), and BD MGIT OADC enrichment supplement (BD, SA) was prepared, and 10 mL was dispensed into a filtered screwcap tissue culture flask 25 cm² (Separations, SA). The culture flask was then inoculated with the laboratory strain Mtb, H37Rv (ATCC 27294) and left to incubate for 2 weeks at 37°C. Following incubation, the culture was transferred to a 15-mL falcon tube, centrifuged at 4,000 rpm for 20 min, and the supernatant was discarded. The pellet was then resuspended in 200 µL lysis buffer (0.05M Tris-HCl, 0.05M EDTA, pH 8) prior to DNA extraction. Following extraction as described below, the DNA was diluted five times to generate samples consisting of approximately 105, 104, 103, 102, and 10 genomes/µL. This was estimated by considering the Mtb genome size of 4.4 Mb, expecting ~4.52 fg of gDNA to correspond to one Mtb genome. The starting sample had DNA concentration of 4.52 ng/µL or ~106 H37Rv genomes and diluted 10-fold from there for each sample.
Sediment sample preparation and pre-treatment to deplete contaminating host DNA
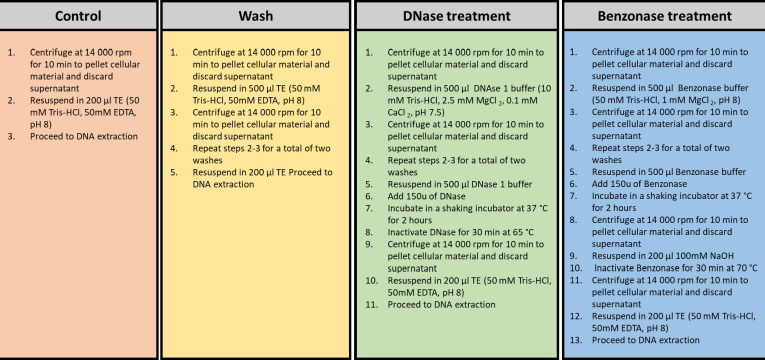
Routinely discarded Xpert positive sediments (n = 8) (NALC-NaOH decontaminated sputum samples, referred to as sediments) were collected from the National Health Laboratory Services Green Point, Western Cape, Cape Town, South Africa. The samples used for this study were remnants from an accompanying sputum sample decontaminated with NALC-NaOH for culture and line probe assay and were thus not exposed to the Xpert sterilizing reagent. Samples were frozen at –20°C until further processing. Sediments were pooled to a final volume of 15 mL, heat inactivated for 1 hour at 80°C, removed from the biosafety level 3 facility, and then aliquoted into 15 × 1 mL aliquots with intermittent mixing between each aliquot. The 15 aliquots were then split into groups of three replicates each per treatment condition namely a control, wash, benzonase treatment, and DNase treatment group. Sample treatment was subsequently processed as depicted in Fig. 1.
Fig 1.
Summary of pre-treatment steps to deplete extracellular contaminating DNA prior to DNA extraction.
DNA extraction
DNA was extracted using the Zymo-DNA Clean & Concentrator-25 (Zymo, USA) according to the manufacturer’s instructions (17) . Briefly, 50 µL of 100 mg/mL of lysozyme (Merck, Germany) was added to each sediment aliquot and incubated overnight at 37°C with gentle mixing. Thereafter, 50 µL of 2.5 mg/mL proteinase K (Merck, Germany) and 100 µL of 20% SDS (Thermofisher Scientific, USA) were added and incubated at 65°C for 30 min. Binding buffer (800 µL) was added and gently mixed by inverting the tube until the sample solidified, then mixed vigorously by hand until the sample returned to a liquid state, and then gently mixed for 5 min. The solution was transferred to the column provided and centrifuged at 14,000 rpm for 30 s, and this step was repeated until all of the solution had been loaded onto the column. The flow through was discarded after each centrifugation step. The column was then washed twice with 200 µL of wash buffer, by adding the wash buffer to the column and centrifugation at 14,000 rpm for 60 s. To elute the purified DNA, 50 µL of preheated elution buffer (10 mM Tris-HCl) is added to the column, incubated for 5 min, and then centrifuged at 14,000 rpm for 30 s. The elution step was then repeated with an additional 50 µL aliquot of heated elution buffer to improve the final yield. The eluted DNA concentrations were quantified using the Qubit HS dsDNA Assay Kit and the Qubit ssDNA Assay Kit (Thermofisher Scientific, USA).
qPCR
Luna Universal qPCR Master Mix (New England Biolabs, USA) and other reaction components were thawed at room temperature and gently vortexed. The final 20 µL volume for each reaction consisted of 10 µL 2X Luna Universal qPCR Master Mix [which contains deoxynucleotide triphosphate (dATP, dTTP, dCTP, and dGTP), MgCl2, Hot Start Taq DNA Polymerase, fluorescent dye, and a passive reference dye], 0.5 µL forward primer (10 µM), 0.5 µL reverse primer (10 µM) for either the the human-specific target PTGER2 (primer pairs; hPTGER2f (5′-GCTGCTTCTCATTGTCTCGG-3′) and PTGER2r (5′-GCCAGGAGAATGAGGTGGTC-3′)) (18), or the Mtb-specific target Rv2341 (primer pairs; Rv2341-F (5′-GCCGCTCATGCTCCTTGGAT-3′) and Rv2341-F (5′-AGGTCGGTTCGCTGGTCTTG-3′)) (19), 5 µL nuclease free water and 4 µL of template (1 ng purified DNA). The “SYBR/FAM” channel of the CFX96 Touch Real-Time PCR Detection System was used for quantification (Biorad, USA). Cycling conditions were as follows: initial denaturation at 95°C for 1 min; 40 cycles consisting of 95°C for 15 s, 62°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 2 min. All PCR amplification reactions were done in triplicate.
qPCR analyses
Ct values were used to determine relative target abundance between samples or to measure absolute target quantities based on an acceptable standard curve obtained from a set of known dilutions. Standard curves were prepared at five concentrations by serial dilution, equating to input amounts of 10, 1, 0.1, 0.01, and 0.001 ng/per reaction of either Mtb or host DNA. The average Ct for each sample was taken and used along with the standard curve to calculate the approximate copy number of either Mtb or host DNA to ascertain the ratio of target to host DNA (Mtb DNA:host DNA). Copy number estimates were normalized per nanogram input DNA.
Library preparation and target capture and enrichment
Twist target capture probes were custom designed for onefold coverage of the full H37Rv genome with duplicates removed, supplemented by fourfold coverage of 95 known lineage single-nucleotide variants (SNVs) (20) and 1,387 homoplasic SNVs and 60 INDELs in drug resistance regions (Supplementary folder 1—Probe panel). Targeted mutations were merged within a 60-bp region and tiled as a block fourfold. Probes were manufactured by Twist Biosciences, San Francisco, USA, and shipped to Stellenbosch University, South Africa.
The limit of detection range as well as one sample from each treatment condition of the benzonase/DNase head-to-head comparison experiment were selected for Twist target capture and sequencing (characteristics for the final pool of samples can be found in Table S5). All eight samples were subjected to library preparation using the Twist enzymatic fragmentation and universal adapter system as follows. Library preparation was done according to the manufacturer’s instructions with only two modifications. Considering the low input concentrations and based upon feedback from the manufacturer, the adapter concentration was halved and the amplification cycles using the Twist UDI primers during the library preparation step were increased from 8 to 10 cycles. The eight prepared libraries were pooled for hybridization and post capture amplification, the resulting library was run on a MiniSeq sequencer (Illumina, USA), using the MiniSeq High Output Reagent Kit (150 cycles).
Bioinformatics
Initial quality assessment was done using fastQC, version 0.11.9, followed by adapter trimming, quality filtering, and per-read quality pruning using fastP, version 0.20.1 (21). Following quality control steps, reads were taxonomically classified using the metagenomic classification tool Kraken2 (version 2.0.8). A custom kraken2 database was built using GenBank/RefSeq sequences and included all GenBank bacteria, fungi, plasmid, viral, and phage sequences (complete, chromosome, and scaffold). The RefSeq human genome along with additional Mtb genomes (CP011510_1, NC_000962_3, NC_002755_2, NC_009565_1, NC_017524_1, NZ_OW052188_1, and NZ_OW052302_1) were also added to the database (22). Reads classified as Mtb were extracted using the KrakenTools extract_kraken_reads.py script. Both filtered and unfiltered fastq files were aligned using bwa-mem2 (version 2.2.1) to the H37Rv reference genome AL123456 (23). Duplicates were removed using Picard tools and excluded in further downstream analyses. Alignment statistics—including number of reads, depth and breadth of coverage, and GC-content—were determined and visualized using Qualimap (version 2.2.2 c). Outputs from Qualimap were used to generate plots of the normalized sequencing depth across the reference genome, and any observed regions with deviations in coverage were visualized using Integrative Genomics Viewer (IGV) (version 2.3.97) (24).
Statistical analyses
Statistically significant differences between control and treatment groups were determined using the unpaired t-test with Welch’s correction (P-value <0.05), for both the DNA concentrations as determined by the Qubit ssDNA and Qubit HS dsDNA assays, as well as for the copy number estimates based on qPCR results.
RESULTS
DNA extraction and quantification: ssDNA vs dsDNA
Average DNA concentrations in treated samples and controls measured by the Qubit ssDNA and Qubit HS dsDNA assays are outlined in Table 1. Comparison of the dsDNA and ssDNA assay results suggests that the HS dsDNA assay underestimates total DNA in the sample after treatment of NALC-NaOH decontaminated samples (Table 1). The dsDNA and ssDNA quantification assays both demonstrated that on average all treatment groups had a lower final DNA concentration following DNA extraction in comparison to the controls and that both DNase and benzonase treatments lead to a significant reduction in the total DNA concentration of each sample. This reduction suggests that there is significant amount of extracellular DNA in decontaminated sputum sediments, and we expect the majority of this DNA to be single stranded given the exposure to high pH conditions.
TABLE 1.
Average DNA concentrations of sample groups as measured by Qubit HS dsDNA assay and Qubit ssDNA assay for the head-to-head comparison a
| Treatment | DNA concentration ng/µL | P-value treatment vs control |
|---|---|---|
| Qubit HS dsDNA assay | ||
| Control | 1.6 ± 0.32 | NA b |
| Wash | 1 ± 0.27 | 0.0491 |
| DNase | 0.37 ± 0.08 | 0.0146 |
| Benzonase | 0.38 ± 0.09 | 0.0151 |
| Qubit ssDNA assay | ||
| Control | 5.32 ± 1.52 | NA |
| Wash | 2.7 ± 1.36 | 0.0911 |
| DNase | 0.55 ± 0.30 | 0.0297 |
| Benzonase | 0.51 ± 0.21 | 0.0283 |
Significant differences between control and treatment groups were determined by unpaired t-test with Welch’s correction (p-value < 0.05). Three technical replicates were done per treatment group.
NA, not applicable.
qPCR quantification of Mtb and host DNA
Analyzing the quantitative PCR results for the control demonstrated a higher average copy number of the host genome (6,168 ± 1,638 copies/ng) relative to the Mtb target (212.3 ± 59.4 copies/ng), with an unfavorable ratio of 1:29 target vs host genomic copies. A simple wash of the sample or its treatment according to the DNase protocol (which includes two washes with DNAse buffer as well as the DNase treatment, Fig. 1) led to a significant improvement in the copy numbers present for Mtb target DNA and a significant reduction in contaminant host DNA, as well as an improved ratio of Mtb DNA:host DNA when comparing the treatment conditions to the control (Table 2).
TABLE 2.
Decontamination by wash vs DNase vs benzonase prior to cell lysis: copy number/ng of input DNA and ratios of Mtb:host are computed from qPCR performed on same starting material that was split fourfold between the control and three test protocols a
| Treatment | Mtb copy number/ng | Mtb treated vs control P value | Host copy number/ng | Host treated vs control P value | Mtb DNA:host DNA ratio |
|---|---|---|---|---|---|
| Control | 212.3 ± 59.4 | NA b | 6,168 ± 1,638 | NA | 1:29 |
| Wash | 3,151 ± 1,369 | 0.0002 | 2,726 ± 936.4 | 0.0001 | 1:0.86 |
| DNase | 2,862 ± 733.4 | 0.0001 | 2,414 ± 1,043 | 0.0001 | 1:0.84 |
| Benzonase | 11,721 ± 7,096 | 0.0012 | 3,422 ± 2,162 | 0.0084 | 1:0.29 |
Summary of Mtb copy number and host copy number per 1 ng of input DNA as determined by qPCR and expressed as a ratio of Mtb to host copy number. Significant differences between control and treatment groups were determined by unpaired t-test with Welch’s correction (P < 0.05). Three technical qPCR replicates were done for every sample.
NA - Not applicable.
No significant difference was observed between either the Mtb copy number (P = 0.6) or the host copy number (P = 0.5), or the ratio of Mtb DNA:host DNA (1:0.86 vs 1:0.84) when directly comparing the washed and DNase-treated samples. This suggests that the impact is mainly due to the wash steps of the cellular material. Benzonase treatment showed enrichment for Mtb DNA, resulting in a 100-fold enrichment in the ratio of Mtb DNA:host DNA using benzonase (1:0.29) over the control (1:29) (Table 1). In summary, a simple wash of the decontaminated sample leads to an improved ratio of Mtb to host DNA, but additional pre-treatment with benzonase resulted in further enrichment of the Mtb target.
Enrichment with Twist target capture
Limit of detection
To initially evaluate the Twist target capture system, we assessed the LoD using a series of samples spiked with H37Rv DNA at progressively lower dilutions. The target capture and enrichment system successfully captured and detected Mtb down to a 100-genome copies. The proportion of the reference genome captured with the custom panel was 99.99% for the entire limit of detection range except for the sample equivalent to 10 genomes. We summarized the amount of data sequenced, raw read counts, read quality, insert size, coverage, duplication and error rates, as well as the proportion of the reference genome covered at 5× and 10× (Table S6). Coverage across the reference genome demonstrated high uniformity of capture (Fig. S2). Genomic windows where coverage deviated from the median (>1.5×) were visually inspected by IGV. The regions where an increase in coverage was observed were mainly attributed to insertion sequences (IS6110, IS1081, and IS1557), while regions exhibiting an overall decrease in coverage were attributed to repeat regions (PE, PPE, and Esx).
Direct sputum samples
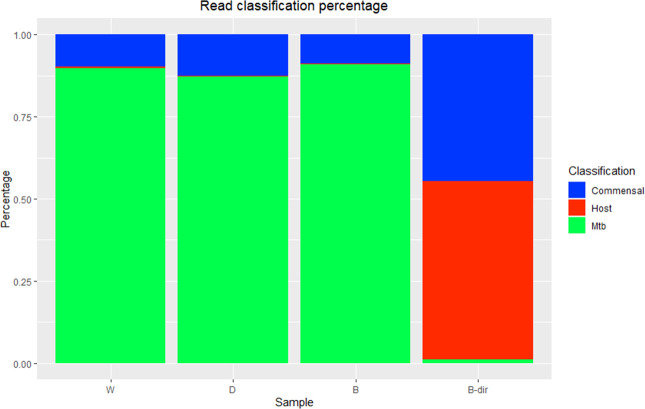
DNA extracted from each of the treatment conditions for the wash vs DNase vs benzonase experiment was included in the Twist target capture system. Classification of reads with kraken2 revealed the percentage of reads classifying as Mtb to be 89.78%, 87.12%, and 90.95% for the wash, DNase-treated, and benzonase-treated samples, respectively (Fig. 2). To assess the effectiveness of enrichment, the benzonase-treated sample was also sequenced directly because it had the highest enrichment ratio by qPCR. For the directly sequenced sample, only 1.18% of reads were classified as target Mtb reads compared to 90.95% for the enriched sample. In addition to the observed enrichment of target-specific reads, 54.24% and 44.58% of reads were classified as belonging to the host or host commensals in the directly sequenced sample compared to 0.32% and 8.73% for the enriched sample. The average depth of coverage obtained for the treated samples was 8× (Wash), 16× (DNase treated), and 22× (Benzonase treated), respectively, and the proportion of the genome covered at least 1× was 99.45% on average with >80% of the genome covered 5× (Table S6). The total amount of data sequenced per sample ranged between 0.05 and 0.13 GB, and the expected coverage per 1 GB sequenced can be found in Table S7.
Fig 2.
Percentage reads classified as either Mtb, host, or other contaminating DNA. W (wash only sample 1—enriched), D (DNase-treated sample 1—enriched), B (Benzonase-treated sample 1—enriched), and B-dir (Benzonase sample 1—unenriched).
Using IGV (25), we visually inspected genomic regions with coverage higher than the median (>1.5×) in the benzonase-treated sample (Fig. S3). Similar to the LoD experiments, an increase in coverage was observed at insertion sequences (IS6110, IS1081, and IS1557), while repeat regions (PE, PPE, and esX) exhibited a decrease in overall coverage. For example, IS6610 elements had an average coverage of 77×, and as a representative example, PE_PGRS4 had an average coverage of 3.8×. There was also a significant increase in the average coverage (891×) observed across the rrs and rrl genes. Several other regions conserved among bacterial species such as clpB, HSP, rpoB, rpoC, tuf, rpsc, and aspT were also found to have more than expected coverage ranging between five- and ninefold higher than the genomic average.
Given the high conservation of the rrs/rrl regions and the aforementioned genes, we hypothesized that the observed high coverage is due to off-target capture of these elements from non-Mtb respiratory bacteria contaminating the sample. We classified all bacterial reads mapping to the rrs-rrl region (13.50% of total sequencing reads) using the Kraken2 (26). Only 4.83% of these reads were classified as Mtb confirming that the variations in coverage were due to off-target capture. The six most abundant source genera for the rrsl/rrl reads overrepresented respiratory flora including Streptococcus (19.68%), Streptomyces (5.62%), Arthrobacter (3.92%), Rhodococcus (1.42%), Bifidobacterium (1.52%), and Staphylococcus (1.63%). We then used Kraken2 to remove contaminating DNA across the whole genome, by excluding all reads not classified as Mtb complex (9.05% of total) prior to mapping to the H37Rv reference genome. This corrected the non-uniformity of coverage in all regions except for 16S (rrs) and 23S (rrl) that were still covered at 2.5-fold across both genes and up to 15-fold the genomic average in certain regions.
DISCUSSION
Sputum specimens collected for Mtb detection remain challenging for DSS due to the large quantities of contaminating host DNA they contain. We aimed to evaluate two approaches, both separately and in conjunction, to facilitate DSS. Results reveal that host DNA can be successfully depleted by washing the samples or treating the samples with DNase or benzonase, leading to a significantly improved ratio of Mtb DNA:host DNA. Ideal results required target capture and enrichment as well, which we included via a DNA probe-based capture system from Twist Bioscience (2 – 4, 6). Finally, we also report that dsDNA assays do not accurately estimate DNA concentrations when working with NaOH decontaminated sediments, underestimating DNA concentration compared to an ssDNA assay.
In standard procedures, sputum samples are decontaminated under alkaline conditions (pH 12–14) prior to downstream processing for Mtb culture (5). Alkaline conditions (pH >9), such as those generated by sodium hydroxide, denature DNA leading to large amounts of ssDNA in the sample. This observation is important since techniques like qPCR and NGS rely on accurate DNA quantification in the input sample, and incorrect quantification could also impact the final results (27, 28). Alkaline conditions and heat inactivation, as used in our protocol, can both lyse contaminating host cells and Gram-negative organisms. This then causes the majority of the contaminating DNA to become extracellular and available for enzymatic degradation (10, 11), meaning washing the sediment sample prior to DNA extraction will remove most of these contaminants. This is congruent with previous studies where the application of a saline wash or treatment with the MolYsis kit (Molzym, Germany) reduced contaminating DNA (5, 29).
The benzonase treatment protocol demonstrated a significant enrichment effect. We initially hypothesized that benzonase was superior to a simple wash or DNase treatment because it more effectively removes extracellular contaminating ssDNA in addition to dsDNA, RNA, and DNA:RNA hybrids, while DNase targets only dsDNA (12 – 14). However, there was only a small difference in total DNA by the Qubit ssDNA between the DNase- and benzonase-treated samples (0.55 ± 0.302 ng/µL and 0.51 ± 0.213 ng/µL, respectively), arguing that more effective ssDNA degradation is not the only reason for the benzonase effect. An alternative explanation is that the remaining extracellular DNA is not accessible to the DNAse and that the benzonase treatment protocol may render the DNA more accessible to the benzonase enzyme. On the other hand, the additional exposure to alkaline conditions can also result in more effective Mtb lysis than the other two protocols, thus enhancing recovery of Mtb. The exact mechanisms behind the observed enrichment effect will be the focus of future studies (11).
Although a positive enrichment effect was observed, target capture and enrichment were still necessary because of the low levels of target DNA from DSS (3 – 5). Our results demonstrate that the Twist kit can handle low input amounts of DNA down to 100-Mtb genome copies (0.45 pg of Mtb input DNA), making it an important breakthrough necessary for success for DSS. After Twist enrichment, WGS quality, expected coverage, and uniformity were high. Sequencing data demonstrated a very low proportion of duplicate reads (median 0.43%) in relation to a previous study which reported an extremely high proportion of duplicates (median 80%) for samples enriched with RNA baits (3). The reduced duplication rate can be in response to the Twist kits unique chemistry, but further research and in-depth analyses of various enrichment kit protocols are warranted to determine the precise steps of the reaction that contribute this.
In addition to the low propensity to capture host DNA and low potential for biased capture, we have demonstrated the effectiveness of the Twist target capture and enrichment system to enrich for the Mtb target from 1.18% in the directly sequenced sample to 90.95% in the enriched. We also found no added benefit with additional pre-treatments prior to use of the Twist kit other than recommended wash steps as the proportion of reads attributed ranged between 87.12% and 90.95% and were sufficient for the recovery of the target genome for all three treatment conditions. We did identify off-target capture (9.05% of total reads) for highly conserved domains, especially the rRNA regions from contaminating respiratory flora, but not from host DNA. The off-target capture of rRNA elements is not unique to the evaluated enrichment platform, and several previous studies utilizing RNA baits have also reported off-target capture and poor uniformity in these regions (2, 4).
Current technologies employed for the target capture and enrichment of Mtb rely on hybridization capture and polymerase chain reaction-based methods to facilitate the capture and enrichment of the target. There is thus a potential for the introduction of sequencing bias in regard to capture efficiency, capture specificity, target coverage, GC and AT dropout, sensitivity to single-nucleotide polymorphisms, and insertions and deletions, all of which may impact final results (30, 31). Previous studies, that applied a target capture approach to Mtb to compare culture to DSS, conclude that DSS captures a higher degree of diversity. The authors did not highlight any potential for the introduction of bias to the population structure in regard to the capture technology employed except for the off-target capture of rRNA elements and have similar to the current study demonstrated good Mtb genome uniformity (2 – 4, 6).
Limitations
The current study has limitations. The LoD experiments consisted of pure H37Rv in culture media and not in sputum and hence excluded the effects of contaminating DNA. This allowed us to isolate the effect of the inoculum and study sequencing uniformity independent of contamination. We also note that the generated LoD was done with DNA quantities and that this does not always correspond to colony forming units (CFUs). For the sputum-based experiments, we used pooled sputum sediments that increased the risk of contamination but was needed to generate enough sample volume for protocol comparisons and technical replicates (32, 33). Individual sediments pooled were not quantified prior to pooling, and for future studies, pools can be generated from various smear grades to generate representative low, moderate, and high Mtb load samples. Due to the limited amount of DNA that can be extracted from this sample type, we opted to only quantify host and target DNA, thus omitting the quantification of overall bacterial load. This decision was made to prioritize host DNA quantification as host DNA is generally the primary contributing contaminant in clinical specimens (2 – 4, 6).
We note that our input samples were generated from a pool of several sediments. There was thus an expected higher risk of contamination and constituted a more extreme test of the specificity of capture compared with a single patient isolate. Off-target capture was readily addressable bioinformatically by excluding non-Mtbc reads. Prior work on DSS also suggested removing or masking the rRNA genes in WGS data prior to variant calling (2, 4, 34). Despite the bioinformatics solution, off-target capture increases cost and decreases efficiency of DSS. Additionally, the genes identified in which off-target capture has been seen, although homologs to some extent, have genetic differences that when captured with higher accuracy are used to specifically distinguish Mtb from other organisms emphasizing the need for improved specificity. In the future, redesign of the panel to exclude or de-enrich these regions may be considered. The probe panel was supplemented by fourfold coverage for homoplastic SNVs in drug resistance regions, 91 of which are found in the rrs and rrl genes, potentially exacerbating the extent of off-target capture. Alternatively, refinement of current protocols to prevent non-specific binding e.g., by capture at higher temperatures, may also reduce off-target capture (35).
Conclusions
We demonstrate that pre-processing with a benzonase enrichment protocol to remove contaminating extracellular DNA prior to cell lysis and DNA extraction has a positive effect for the enrichment of Mtb DNA in decontaminated sediments, but the exact mechanism by which enrichment is facilitated is still unknown. In addition, we demonstrate the utility and low limit of detection of the DNA probe-based Twist target capture and enrichment system for enrichment and sequencing of Mtb from clinical sputum sediments. The reported Twist target capture and enrichment system shows promise for future clinical application to enable direct sputum sequencing. Direct sputum sequencing can be initiated after GeneXpert-MTB/RIF confirms the presence of Mtb, to characterize bacterial diversity with higher degree of sensitivity than culture, and by capturing the whole genome. This can assist with drug resistance profiling and identifying resistance markers that may be missed with standard targeted molecular diagnostic tests.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH-R01 AI155765). We would also like to thank Y.G. and the National Health Laboratory Services (NHLS), TB Molecular Laboratory, Green Point, for assistance with sample collection as well as the South African Medical Research Council (SAMRC) for utilization of their facilities where the MiniSeq used during this study is housed. All claims expressed in this article are solely those of the authors and do not represent those of their affiliated organizations, or the funders. The funder had no role in study design, data, data analysis, the decision to publish, or preparation of the manuscript.
B.C.M., M.R.F., and R.M.W. designed the study with key contributions from K.R.J. B.C.M. preformed the sample preparation, DNA extraction, sequencing, and the data analysis with key input from M.R.F and R.M.W. B.C.M. wrote the first draft of the manuscript and M.R.F., R.M.W., K.R.J., and Y.G. provided key edits of the manuscript.
Contributor Information
B. C. Mann, Email: bcmann@sun.ac.za.
Melissa B. Miller, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, North Carolina, USA
DATA AVAILABILITY
The MiniSeq data obtained in this study have been deposited in the NCBI Sequence Read Archive under project accession number PRJNA987443.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jcm.00382-23.
Dataset S1 - AR_homoplasic_INDELs.
Dataset S2 - AR_homoplasic_SNVs.
File S1 - Lineage_barcode.
Standard curve generated to validate dilution accuracy of the generated H37Rv limit of detection.
Normalized sequencing depth across the reference genome using different input amounts of target DNA for all H37Rv LoD samples.
Normalized sequencing depth across the reference genome for all enriched sediment samples (B - Benzonase treated, D - DNase treated and W - Washed).
File S2 - probe_placement_Farhat_MTuberculosis_NC_000962_3_AR_Lineage_TE-93871834_38813.
Supplementary document containing all supplementary tables, figures and file headings.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Dheda K, Perumal T, Moultrie H, Perumal R, Esmail A, Scott AJ, Udwadia Z, Chang KC, Peter J, Pooran A, von Delft A, von Delft D, Martinson N, Loveday M, Charalambous S, Kachingwe E, Jassat W, Cohen C, Tempia S, Fennelly K, Pai M. 2022. The intersecting pandemics of tuberculosis and COVID-19: population-level and patient-level impact, clinical presentation, and corrective interventions. Lancet Respir Med 10:603–622. doi: 10.1016/S2213-2600(22)00092-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown AC, Bryant JM, Einer-Jensen K, Holdstock J, Houniet DT, Chan JZM, Depledge DP, Nikolayevskyy V, Broda A, Stone MJ, Christiansen MT, Williams R, McAndrew MB, Tutill H, Brown J, Melzer M, Rosmarin C, McHugh TD, Shorten RJ, Drobniewski F, Speight G, Breuer J. 2015. Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples. J Clin Microbiol 53:2230–2237. doi: 10.1128/JCM.00486-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goig GA, Cancino-Muñoz I, Torres-Puente M, Villamayor LM, Navarro D, Borrás R, Comas I. 2020. Whole-genome sequencing of Mycobacterium tuberculosis directly from clinical samples for high-resolution genomic epidemiology and drug resistance surveillance: an observational study. Lancet Microbe 1:e175–e183. doi: 10.1016/S2666-5247(20)30060-4 [DOI] [PubMed] [Google Scholar]
- 4. Nimmo C, Shaw LP, Doyle R, Williams R, Brien K, Burgess C, Breuer J, Balloux F, Pym AS. 2019. Whole genome sequencing Mycobacterium tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture. BMC Genomics 20:433. doi: 10.1186/s12864-019-5841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Votintseva AA, Bradley P, Pankhurst L, Del Ojo Elias C, Loose M, Nilgiriwala K, Chatterjee A, Smith EG, Sanderson N, Walker TM, Morgan MR, Wyllie DH, Walker AS, Peto TEA, Crook DW, Iqbal Z. 2017. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 55:1285–1298. doi: 10.1128/JCM.02483-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doyle RM, Burgess C, Williams R, Gorton R, Booth H, Brown J, Bryant JM, Chan J, Creer D, Holdstock J, Kunst H, Lozewicz S, Platt G, Romero EY, Speight G, Tiberi S, Abubakar I, Lipman M, McHugh TD, Breuer J. 2018. Direct whole-genome sequencing of sputum accurately identifies drug-resistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J Clin Microbiol 56:e00666-18. doi: 10.1128/JCM.00666-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soundararajan L, Kambli P, Priyadarshini S, Let B, Murugan S, Iravatham C, Tornheim JA, Rodrigues C, Gupta R, Ramprasad VL. 2020. Whole genome enrichment approach for rapid detection of Mycobacterium tuberculosis and drug resistance-associated mutations from direct sputum sequencing. Tuberculosis (Edinb) 121:101915. doi: 10.1016/j.tube.2020.101915 [DOI] [PubMed] [Google Scholar]
- 8. Bodi K, Perera AG, Adams PS, Bintzler D, Dewar K, Grove DS, Kieleczawa J, Lyons RH, Neubert TA, Noll AC, Singh S, Steen R, Zianni M. 2013. Comparison of commercially available target enrichment methods for next-generation sequencing. J Biomol Tech 24:73–86. doi: 10.7171/jbt.13-2402-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kozarewa I, Armisen J, Gardner AF, Slatko BE, Hendrickson CL. 2015. Overview of target enrichment strategies. Curr Protoc Mol Biol 112:7. doi: 10.1002/0471142727.mb0721s112 [DOI] [PubMed] [Google Scholar]
- 10. Hasan MR, Rawat A, Tang P, Jithesh PV, Thomas E, Tan R, Tilley P. 2016. Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J Clin Microbiol 54:919–927. doi: 10.1128/JCM.03050-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shehadul Islam M, Aryasomayajula A, Selvaganapathy PR. 2017. A review on macroscale and microscale cell lysis methods. Micromachines 8:83. doi: 10.3390/mi8030083 [DOI] [Google Scholar]
- 12. Sutton DH, Conn GL, Brown T, Lane AN. 1997. The dependence of DNase I activity on the conformation of oligodeoxynucleotides. Biochem J 321:481–486. doi: 10.1042/bj3210481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu J, Li Z, Li J, Liu Z. 2019. Application of benzonase in preparation of decellularized lamellar porcine corneal stroma for lamellar keratoplasty. J Biomed Mater Res A 107:2547–2555. doi: 10.1002/jbm.a.36760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amar Y, Lagkouvardos I, Silva RL, Ishola OA, Foesel BU, Kublik S, Schöler A, Niedermeier S, Bleuel R, Zink A, Neuhaus K, Schloter M, Biedermann T, Köberle M. 2021. Pre-digest of unprotected DNA by Benzonase improves the representation of living skin bacteria and efficiently depletes host DNA. Microbiome 9:123. doi: 10.1186/s40168-021-01067-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagy-Szakal D, Couto-Rodriguez M, Wells HL, Barrows JE, Debieu M, Butcher K, Chen S, Berki A, Hager C, Boorstein RJ, Taylor MK, Jonsson CB, Mason CE, O’Hara NB. 2021. Targeted hybridization capture of SARS-CoV-2 and metagenomics enables genetic variant discovery and nasal microbiome insights. Microbiol Spectr 9:e0019721. doi: 10.1128/Spectrum.00197-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim KW, Deveson IW, Pang CNI, Yeang M, Naing Z, Adikari T, Hammond JM, Stevanovski I, Beukers AG, Verich A, Yin S, McFarlane D, Wilkins MR, Stelzer-Braid S, Bull RA, Craig ME, van Hal SJ, Rawlinson WD. 2021. Respiratory viral co-infections among SARS-CoV-2 cases confirmed by virome capture sequencing. Sci Rep 11:3934. doi: 10.1038/s41598-021-83642-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Epperson LE, Strong M. 2020. A Scalable, efficient, and safe method to prepare high quality DNA from mycobacteria and other challenging cells. Journal of clinical tuberculosis and other mycobacterial diseases 19:100150. doi: 10.1016/j.jctube.2020.100150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marotz CA, Sanders JG, Zuniga C, Zaramela LS, Knight R, Zengler K. 2018. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome 6:42. doi: 10.1186/s40168-018-0426-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goig GA, Torres-Puente M, Mariner-Llicer C, Villamayor LM, Chiner-Oms Á, Gil-Brusola A, Borrás R, Comas Espadas I. 2020. Towards next-generation diagnostics for tuberculosis: identification of novel molecular targets by large-scale comparative genomics. Bioinformatics 36:985–989. doi: 10.1093/bioinformatics/btz729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freschi L, Vargas R, Husain A, Kamal SMM, Skrahina A, Tahseen S, Ismail N, Barbova A, Niemann S, Cirillo DM, Dean AS, Zignol M, Farhat MR. 2021. Population structure, biogeography and transmissibility of Mycobacterium tuberculosis. Nat Commun 12:6099. doi: 10.1038/s41467-021-26248-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vasimuddin M, Misra S, Li H, Aluru S. 2019. Efficient architecture-aware acceleration of BWA-MEM for Multicore systems, p 314–324. In 2019 IEEE International parallel and distributed processing symposium (IPDPS. IEEE, Rio de Janeiro, Brazil. doi: 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
- 24. Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu J, Rincon N, Wood DE, Breitwieser FP, Pockrandt C, Langmead B, Salzberg SL, Steinegger M. 2022. Metagenome analysis using the Kraken software suite. Nat Protoc 17:2815–2839. doi: 10.1038/s41596-022-00738-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X, Lim HJ, Son A. 2014. Characterization of denaturation and renaturation of DNA for DNA hybridization. Environ Health Toxicol 29:e2014007. doi: 10.5620/eht.2014.29.e2014007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He H-J, Stein EV, DeRose P, Cole KD. 2018. Limitations of methods for measuring the concentration of human genomic DNA and oligonucleotide samples. Biotechniques 64:59–68. doi: 10.2144/btn-2017-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Votintseva AA, Pankhurst LJ, Anson LW, Morgan MR, Gascoyne-Binzi D, Walker TM, Quan TP, Wyllie DH, Del Ojo Elias C, Wilcox M, Walker AS, Peto TEA, Crook DW. 2015. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol 53:1137–1143. doi: 10.1128/JCM.03073-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andermann T, Torres Jiménez MF, Matos-Maraví P, Batista R, Blanco-Pastor JL, Gustafsson ALS, Kistler L, Liberal IM, Oxelman B, Bacon CD, Antonelli A. 2019. A guide to carrying out a phylogenomic target sequence capture project. Front Genet 10:1407. doi: 10.3389/fgene.2019.01407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou J, Zhang M, Li X, Wang Z, Pan D, Shi Y. 2021. Performance comparison of four types of target enrichment baits for exome DNA sequencing. Hereditas 158:10. doi: 10.1186/s41065-021-00171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burdz TVN, Wolfe J, Kabani A. 2003. Evaluation of sputum decontamination methods for Mycobacterium tuberculosis using viable colony counts and flow cytometry. Diagn Microbiol Infect Dis 47:503–509. doi: 10.1016/s0732-8893(03)00138-x [DOI] [PubMed] [Google Scholar]
- 33. Asmar S, Drancourt M. 2015. Chlorhexidine decontamination of sputum for culturing Mycobacterium tuberculosis. BMC Microbiol 15:155. doi: 10.1186/s12866-015-0479-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. George S, Xu Y, Rodger G, Morgan M, Sanderson ND, Hoosdally SJ, Thulborn S, Robinson E, Rathod P, Walker AS, Peto TEA, Crook DW, Dingle KE. 2020. DNA thermo-protection facilitates whole-genome sequencing of mycobacteria direct from clinical samples. J Clin Microbiol 58:e00670-20. doi: 10.1128/JCM.00670-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paijmans JLA, Fickel J, Courtiol A, Hofreiter M, Förster DW. 2016. Impact of enrichment conditions on cross-species capture of fresh and degraded DNA. Mol Ecol Resour 16:42–55. doi: 10.1111/1755-0998.12420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 - AR_homoplasic_INDELs.
Dataset S2 - AR_homoplasic_SNVs.
File S1 - Lineage_barcode.
Standard curve generated to validate dilution accuracy of the generated H37Rv limit of detection.
Normalized sequencing depth across the reference genome using different input amounts of target DNA for all H37Rv LoD samples.
Normalized sequencing depth across the reference genome for all enriched sediment samples (B - Benzonase treated, D - DNase treated and W - Washed).
File S2 - probe_placement_Farhat_MTuberculosis_NC_000962_3_AR_Lineage_TE-93871834_38813.
Supplementary document containing all supplementary tables, figures and file headings.
Data Availability Statement
The MiniSeq data obtained in this study have been deposited in the NCBI Sequence Read Archive under project accession number PRJNA987443.