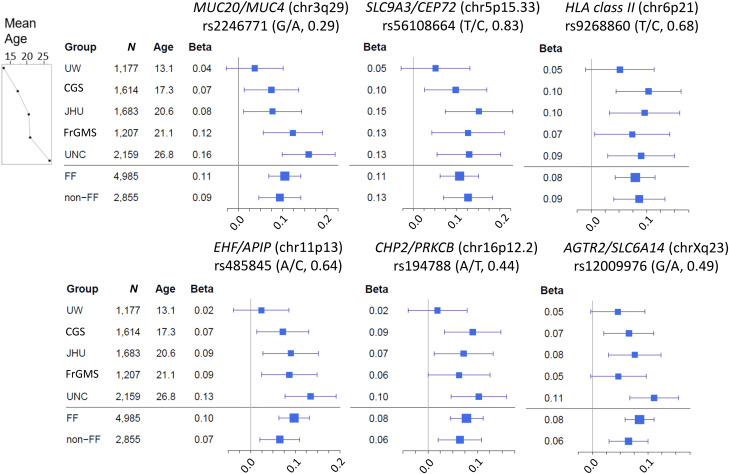
Figure 3.
Forest plots for SNP association effect size by cohort at significant loci. β (coefficient) refers to the average change in Kulich normal residual mortality-adjusted (KNoRMA) phenotype for each copy of the protective allele. Square sizes are proportional to the sample size (n) of each cohort, and the line segments are 95% confidence intervals of each β. The most significant SNP from each locus was chosen. For each SNP, the protective allele is listed on the left, and frequencies of the protective alleles are shown in parentheses. Cohorts are arranged by increasing mean age (at KNoRMA). CFGP = Cystic Fibrosis Genome Project; CGS = Canadian CF Gene Modifier Study (population-based); FF = patients with CF who are F508del homozygous in the CFTR gene; FrGMS = French CF Gene Modifier Consortium (population-based); JHU = Johns Hopkins University (twin-siblings design); non-FF = patients with CF who are not homozygous for F508del in the CFTR gene; UNC = University of North Carolina (extremes of phenotype); UW = University of Washington (longitudinal study for effect of Pseudomonas aeruginosa acquisition on lung disease).