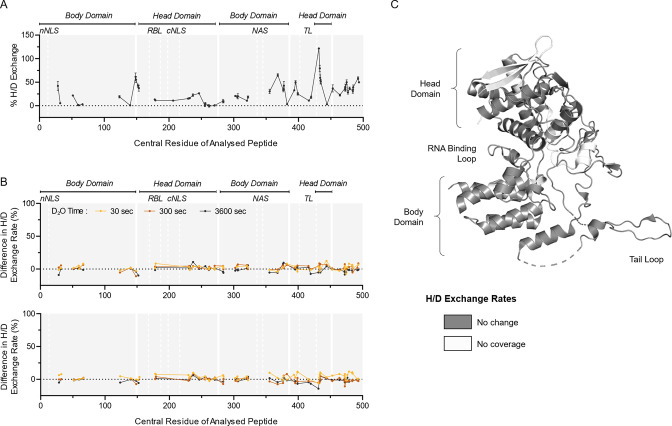
Fig 5.
HDX-MS characterization of NP protein from whole IAV after transient exposure to acidic conditions. (A) H/D exchange percentages were measured for NP protein from whole IAV samples held at neutral conditions (pH 7) for 30 s. Samples were then incubated in D2O for 30 s, quenched, and digested prior to HDX-MS analysis. Data points indicate the mean percentage of H/D exchange ± SD for each peptide plotted using its central amino acid (n = 3, Uniprot: P03466). Continuous lines indicate regions of continuous peptide coverage by MS. Protein domains of interest are indicated by gray shading and white lines; nNLS, nonconventional-conventional nuclear localization signal; RBL, RNA binding loop; cNLS, conventional nuclear localization signal; NAS, nuclear accumulation signal; TL, tail loop. (B) Whole-virus samples were held at neutral pH or adjusted to pH 4, and then neutralized back to pH 7 after 10 or 30 s prior to processing for HDX-MS. Data presented as residual plots showing the difference in H/D exchange percentages induced by exposure to pH 4 when compared to corresponding pH 7 control samples. Individual data points indicate the mean percentage of H/D exchange difference for triplicate samples. (C) Crystal structure of A/WSN/33 NP (PDB: 2IQH), showing regions of coverage by HDX-MS. White indicates regions of no peptide coverage by HDX-MS. Head and body domains, along with RNA binding loop and tail loop of NP, are annotated based on Ye et al. (54).